-
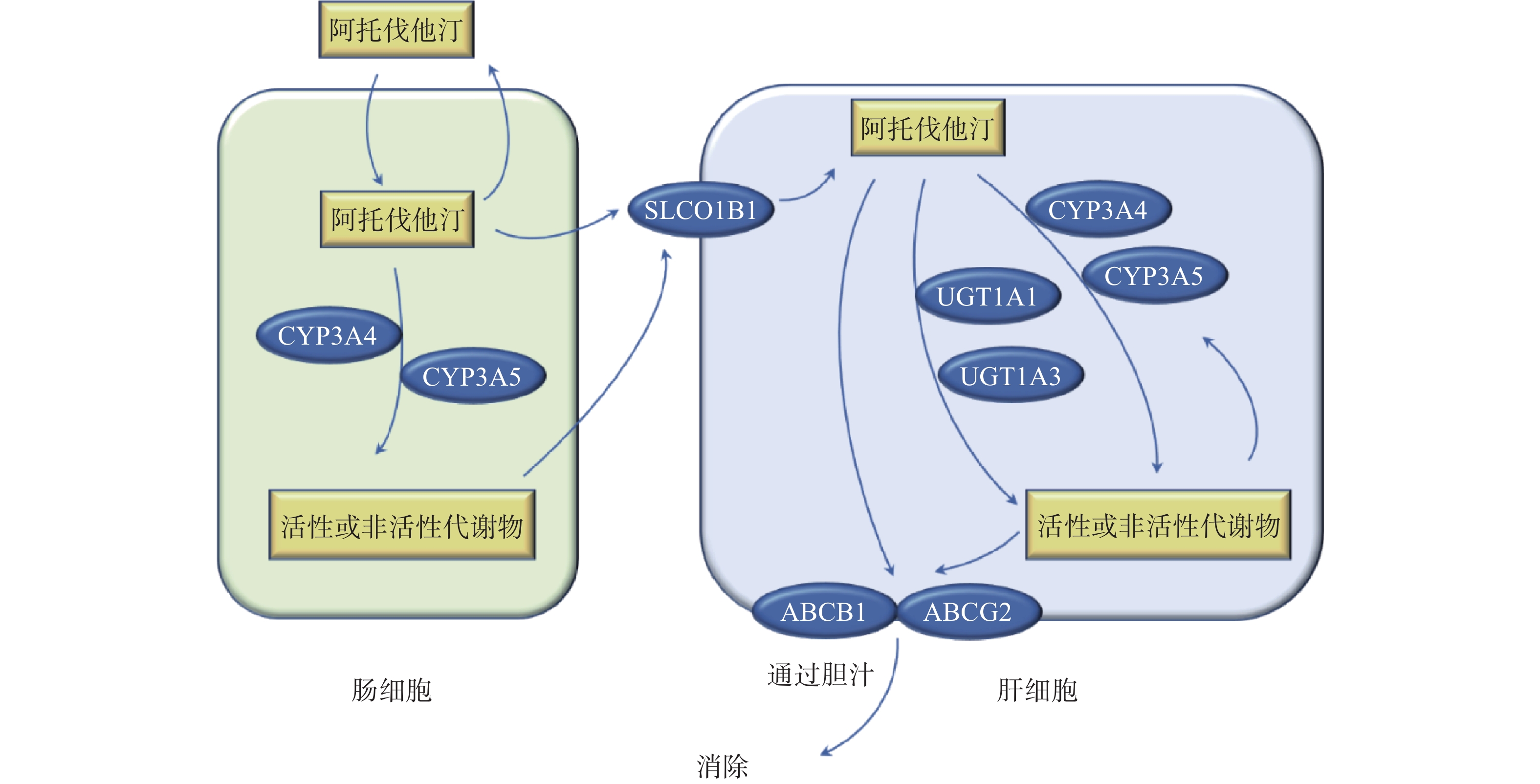
他汀类药物抑制胆固醇合成途径的限速酶3-羟基-3-甲基戊二酸单酰辅酶A还原酶(HMG-CoA还原酶)的活性,抑制胆固醇的生成,上调肝细胞表面的LDL受体以加快LDL的分解代谢。阿托伐他汀以活性酸形式给药,通过被动扩散或OATP1B1转运进入肝细胞,主要由CYP3A4在肝脏和肠壁代谢阿托伐他汀,少量由CYP2C9、CYP2C19、CYP3A5和UGT1A1代谢。阿托伐他汀酸由CYP3A代谢产生邻、对位羟基活性代谢产物,在体内它们与各自的非活性内酯形式处于平衡状态。ABCB1(编码p-gp)和ABCG2(编码BCRP)介导阿托伐他汀及其代谢物从肝脏排入胆汁后消除(图1)[1]。
-
大多数CYP3A代谢发生在肝细胞内,部分发生在小肠内。CYP3A的活性存在较大的个体间差异,显著影响阿托伐他汀的体内代谢。阿托伐他汀和阿托伐他汀内酯主要由CYP3A4同工酶代谢,CYP3A4的活性在不同个体之间可能存在10倍的差异,这可能是由于编码酶的基因多态性所致[2]。
CYP3A4*1G/*1G基因型患者的阿托伐他汀AUC0-∞比野生型和*1/*1G基因型低36%和25%。阿托伐他汀和2-羟基阿托伐他汀的tmax和(CL/Fm)在CYP3A4*1G/*1G基因型和野生型之间存在显著性差异[3]。
经阿托伐他汀治疗后,CYP3A4启动子变异(rs2740574)与野生型等位基因相比显示出更低的LDL-C水平和更好的疗效[2]。CYP3A4*1G(20230G>A)多态性对血清TC下降具有剂量依赖性(P<0.01)。携带CYP3A4*1G增加了阿托伐他汀的降脂效果,对辛伐他汀降脂治疗没有显著影响[4]。CYP3A4*22(rs35599367)会导致CYP3A4酶含量和活性显著降低,CYP3A4*22携带者的阿托伐他汀、辛伐他汀或洛伐他汀的剂量仅为野生型的20%~60%[5]。携带CYP3A4*1B(290A>G)G等位基因可降低辛伐他汀和阿托伐他汀的血药浓度,特别是女性和具有ABCB1 3435T变异等位基因的患者[6]。CYP3A4 A290G变异等位基因与阿托伐他汀治疗后较高的LDL-C水平显著相关,而非同义多态性M445T变异与治疗后较低的LDL-C水平显著相关[7]。此外,携带CYP3A4 A290G变异等位基因比野生型等位基因型的患者具有更高的HDL水平(P<0.001),可能与体内CYP3A4活性降低相关,导致阿托伐他汀活性增加[8]。临床上可减少CYP3A4*22携带者的药物剂量,CYP3A4*1B突变对他汀类药物降脂作用影响尚无定论。
-
CYP3A4和CYP3A5约占CYP450的30%,CYP3A4和CYP3A5均在肝脏和肠道中表达,其中CYP3A5主要在肝外组织中表达。CYP3A5基因与CYP3A家族其他成员一起位于7q22.1染色体上,由9个外显子组成,编码502个氨基酸。
Kivistö等[9]研究发现洛伐他汀、辛伐他汀和阿托伐他汀对CYP3A5表达者(CYP3A5*1)的疗效显著低于非表达者(CYP3A5*3)。携带CYP3A5*1等位基因的受试者1年内的平均血清TC水平和LDL-C水平分别比CYP3A5*3纯合等位基因的受试者高23%(P=0.0014)和24%(P=0.036)。在服用不依赖于CYP3A5代谢的氟伐他汀、普伐他汀的23名受试者中,未发现降血脂与CYP3A5多态性的关联。因此,CYP3A5*3突变可导致CYP3A5酶的表达降低,提高了他汀类药物的降脂疗效。
-
在192例由阿托伐他汀治疗缺血性卒中的中国患者中,Chen等[10]发现CYP2D6 rs1065852显著影响ΔLDL(P<0.001),ΔLDL/LDL (P<0.001),Δ(LDL/HDL)(P<0.001),Δ(LDL/HDL)/(LDL/HDL)(P<0.001)水平。阿托伐他汀对CYP2D6 rs1065852 G等位基因携带者具有较好的降脂效果。
-
阿托伐他汀由UGT1A1和UGT1A3介导的葡萄糖醛酸化转化为相应的内酯形式,UGT1A3是内酯化的主要酶。
Sung等[11]研究表明UGT1A3*2携带者内酯的生成量显著增加。UGT1A3*2/*2组阿托伐他汀内酯和2-羟基阿托伐他汀内酯的AUC分别比UGT1A3*1/*1组高72%和160%。UGT1A3*2携带者的TC和LDL-C降低百分比分别比非携带者低29%和18%。UGT1A3*2多态性与阿托伐他汀内酯化增加相关,可能影响其降脂作用。低表达等位基因UGT1A1*28(TA)7的携带者较正常活性等位基因(TA)6携带者的阿托伐他汀内酯水平更低(P<0.05)[12]。
-
阿托伐他汀易于口服吸收,其吸收取决于转运蛋白的活性,包括摄取性转运体(如有机阴离子转运多肽OATPs)和外排性转运体(乳腺癌耐药蛋白BCRP和P-gp)。阿托伐他汀通过OATP1B1摄取进入肝细胞,通过ABC转运体家族成员(BCRP、P-gp)排泄到胆汁。
-
有机阴离子转运多肽1B1(OATP1B1)表达于人肝细胞的窦状膜上,负责他汀类药物的肝脏摄取,SLCO1B1基因编码。SLCO1B1的SNP影响他汀类药物的体内分布和效能,目前研究最多的两个SNPs为SLCO1B1 521T>C(rs4149056)和388A>G(rs2306283)。
在携带SLCO1B1和ABCG2野生型等位基因的中国和日本受试者的瑞舒伐他汀、阿托伐他汀和辛伐他汀的AUC比白种人更高,其中,阿托伐他汀AUC0-∞分别比白种人高53%和69%[13]。SLCO1B1 c.521CC基因型患者的阿托伐他汀AUC0-48比TT基因型高144%(P<0.001),比TC基因型高61%(P=0.049)[14]。521C等位基因携带者的阿托伐他汀和2-羟基阿托伐他汀的平均Cmax、AUC0-24h和AUC0-∞显著高于521TT基因型组,平均CL值较521TT基因型受试者低,521C等位基因受试者的阿托伐他汀肌毒性风险增加[15]。SLCO1B1 521T>C在阿托伐他汀的临床试验中均可增高AUC,388A>G多态性对疗效影响不显著。
一项体外研究表明[16],SLCO1B1 521T>C下调质膜上OATP1B1的表达以减少阿托伐他汀转运进肝细胞。SLCO1B1 521T>C通过降低肝脏对阿托伐他汀的摄取使药物对HMG-COA还原酶的抑制作用减弱,同时血药浓度升高。SLCO1B1 521C(rs4149056)与辛伐他汀、阿托伐他汀、洛伐他汀和普伐他汀的降脂作用减弱有关[17]。然而,Prado等[18]研究表明,阿托伐他汀对智利高胆固醇血症受试者的降脂作用与SLCO1B1 rs4149056和rs2306283多态性无关(P>0.05),但388A>G rs2306283与阿托伐他汀治疗后更高的HDL-C相关(P=0.02)。现有的研究支持 SLCO1B1 521T>C基因多态性可增加阿托伐他汀的血药浓度,携带突变基因型的患者应适量减少用药剂量。
-
乳腺癌耐药蛋白(BCRP)由ABCG2基因编码,BCRP在肝脏、小肠、胆管等多种正常组织和癌细胞中均有表达。
ABCG2 c.421C>A在非洲人群中突变频率较低,但在白种人中突变频率为10%~14%,在日本或中国受试者中突变频率高达35%[19]。ABCG2 c.421C>A显著影响阿托伐他汀和瑞舒伐他汀的药动学,ABCG2在抑制这类他汀药物肠道吸收方面起到重要作用。c.421AA基因型携带者的瑞舒伐他汀AUC0-∞分别比c.421CA和c.421CC基因型高100%和144%。c.421AA基因型携带者的阿托伐他汀AUC0-∞比c.421CC高72%[1]。携带ABCG2 rs2622604的日本患者的阿托伐他汀口服生物利用度比非携带者增加了55%(95%CI:16%~131%)[20]。亚洲人群的ABCG2 c.421C>A突变率较高,具有变异等位基因的患者使用他汀类药物需减少剂量。
-
MDR1中的同义单核苷酸多态性C3435T和非同义单核苷酸多态性G2677T/A是药物处置和疗效差异的潜在因素。ABCB1 TTT/TTT基因型携带者的辛伐他汀酸AUC0-12h和阿托伐他汀AUC0-∞分别比CGC-CGC高60%和55%,且TTT/TTT型的阿托伐他汀的半衰期比CGC-CGC型长24%(P<0.05)[21]。ABCB1 rs1128503 (1236C>T)、rs2032582 (2677G>T/A)和rs1045642 (3435C>T)显著影响阿托伐他汀的AUC和阿托伐他汀治疗的降脂效果[22]。
ABCB1 C3435T与阿托伐他汀疗效下降相关。Kajinami等[23]发现 C3435T多态性与降LDL-C作用减弱相关,并且仅在女性中存在变异基因型携带者升HDL-C作用增强的现象。Rodrigues等[24]评估了G2677T/A和C3435T MDR1多态性对阿托伐他汀治疗前后血脂水平的影响,阿托伐他汀疗效与MDR1多态性之间没有显著相关性(P>0.05)。单倍型分析显示,与非T/T基因携带者相比,T/T携带者有较高的基础TC和LDL-C水平,提示MDR1多态性可能对巴西高胆固醇血症患者的基础胆固醇水平有重要影响。DeGorter等[25]报道的一项病例对照研究中,未发现ABCB1 2677T与阿托伐他汀血药浓度之间的关联。由于ABCB1变异的临床研究结果不确定且不一致,目前不建议临床常规使用ABCB1基因分型来预测他汀类药物的毒性。尽管如此,ABCB1在他汀类药物转运中发挥重要作用,未来可能包括ABCB1变异的多基因指导他汀类药物治疗。
-
HMGCR是内源性胆固醇合成途径中的限速酶,他汀类药物竞争性抑制HMGCR。外显子13上选择性剪切HMGCR pre-mRNA,产生两个全长(FL)HMGCR和D13 HMGCR[26]。缺少外显子13的选择性剪接HMGCR转录物的表达量与体内TC、LDL-C、APOB和TC的降低呈负相关[27]。Yu等[28]的研究表明,HNRNPA1的过表达降低了HMGCR酶的活性,增强了LDL的摄取并增加APOB。同时,rs1920045是一种与HNRNPA1外显子8选择性剪接相关的SNP,其与他汀类药物降TC作用减弱有关。Leduc等[29]发现HMGCR的选择性剪接可以解释家族性高胆固醇血症患者对他汀类药物应答的22%~55%的差异。携带rs3846662 AA基因型女性在接受他汀治疗后LDL-C的降低百分比显著低于非AA基因型女性(38.4%vs46.2%,P<0.05)。rs3846662多态性和HMGCR mRNA的选择性剪接显著影响女性对他汀治疗的反应。Cuevas等[30]发现在接受阿托伐他汀治疗的智利患者中,HMGCR rs17671591 T等位基因比野生型C等位基因,显示出更明显地降低LDL-C和升高HDL-C效应(LDL降低8%,P=0.021;HDL增加7.7%,P=0.039)[22]。
-
载脂蛋白E (ApoE)是脂质代谢的重要组成部分。ApoE基因的多态性与血脂水平和他汀类药物的降脂反应有关。APOE ε2等位基因的男性携带者LDL-C的降低率比野生型个体高7%~10% (P=0.01)[31]。Cerda等[32]报道APOE ε2等位基因者对高胆固醇血症具有保护作用,并减少致动脉粥样硬化的脂质分布。Deshmukh等[33]发现 APOE rs445925变异与阿托伐他汀治疗后LDL降低相关,而APOE rs4420638变异对阿托伐他汀治疗后LDL下降作用减弱相关。Jenny等[34]报道阿托伐他汀对139名智利受试者的降脂作用与APOE变异相关,E3/4基因型携带者的降胆固醇作用较E3/3基因型差(LDL-C:-18%vs-29%,P<0.001)。E2等位基因携带者的疗效优于E3和E4携带者,E4基因携带者需增加用药剂量才能达到目标调脂作用。
-
CYP7A1为胆固醇7a羟化酶,是催化胆固醇合成胆汁酸排泄途径的限速酶。CYP7A1基因是胆固醇排泄途径中最重要的调控基因,CYP7A1的基因突变可能会影响他汀类药物的反应。rs8192870位于CYP7A1基因第一个内含子,与阿托伐他汀降低LDL有关,阿托伐他汀治疗后,GG/GA基因型携带者LDL水平降低27.89%,AA基因型携带者LDL水平降低35.26%(P=0.021)[35]。
-
ABCG5/G8的胆固醇排泄和CYP7A1催化的胆汁酸生物合成是胆固醇排泄到胆汁中的主要途径。ABCG8 D19H变异体与阿托伐他汀治疗后降低LDL-C作用增强相关,而降TC作用没有显著性差异[36]。ABCG8 H19等位基因携带者比野生型D19等位基因携带者降低LDL-C作用更显著。ABCG8 H19和CYP7A1 C-204 等位基因对阿托伐他汀反应以剂量依赖性方式相互作用[37]。这些组合多态性分析比单一多态性分析(CYP7A1为4.2%,ABCG8 D19H为3.0%)更能有效解释降LDL-C差异,临床上应组合多态性分析以指导特定基因型患者合理用药。
-
胆固醇酯转运蛋白CETP虽然不是他汀类药物的药物靶点,但在胆固醇逆向转运过程、脂蛋白的分解代谢、不同脂蛋白类型的转化中发挥重要作用。CETP 629C/A的突变频率为0.412。与CC或CA基因型携带者相比,AA基因型携带者具有较低的CETP水平(P=0.026)和较高的HDL-C水平(P=0.035)。连续服用阿托伐他汀一年后,CC基因型携带者比CA或AA基因型降低LDL-C、降低LP(A)(P=0.005),升高HDL-C(P=0.045)疗效更显著(P<0.001)[38]。
-
他汀类药物是临床上首选的调血脂药物,并不是所有患者对他汀类药物反应良好,多数患者并未达到调脂目标值,还有部分患者出现不良反应(肌毒性、横纹肌溶解、肝转氨酶持续升高等)。
-
他汀相关肌肉症状(SAMS)是最常见的药物不良反应,常导致患者用药依从性差或停药。他汀类肌炎的特征是肌肉组织炎症导致肌肉疼痛或无力,并伴有肌酸激酶浓度持续升高。SAMS症状出现在开始他汀类药物治疗的最初6个月内,并在他汀类药物剂量降低或停止后消退,他汀类药物治疗需要密切监测血清肌酸激酶(CK)水平以预防SAMS。临床应用阿托伐他汀的群体中,SAMS的发病率更高。欧洲动脉粥样硬化协会共识小组发布的注册和观察性研究中,SAMS的发生率为7%~29%[39]。他汀诱导的肌毒性在体内外均呈现浓度依赖性,从轻度肌痛到潜在致命的横纹肌溶解或暴发性横纹肌溶解合并肌红蛋白尿最终导致急性肾功能衰竭。使用辛伐他汀的患者发生SAMS频率最高,其次是阿托伐他汀,氟伐他汀SAMS频率最低[40]。
遗传变异和高血药浓度是他汀类药物诱导肌病的关键原因。一项涉及1550名阿托伐他汀使用者的Meta分析表明,SLCO1B1 c.521T>C与阿托伐他汀不良反应之间存在显著相关性。未发现SLCO1B1 c.388A>G多态性与不良反应或疗效之间的关联(P>0.05)[41]。SLCO1B1 rs4149056与辛伐他汀治疗后肌病的发生显著相关,辛伐他汀诱导相关肌病的作用可能比阿托伐他汀更强[42]。与SLCO1B1 521 TT基因型携带者相比,TC或CC基因型阿托伐他汀的不良反应发生率高2.3倍[43]。Liu等[44]发现携带至少一个SLCO1B1 521C等位基因的患者发生肌毒性的风险更高。在接受瑞舒伐他汀治疗的个体中,SLCO1B1 521C突变等位基因突变与肌肉毒性风险显著相关。然而,521C突变等位基因与阿托伐他汀和辛伐他汀诱导肌毒性风险之间无显著相关性。SLCO1B1 c.521 T>C突变能降低OATP1B1的转运功能,使他汀类药物血药浓度升高,携带突变基因型的患者应降低药物剂量以减少不良反应。
CYP3A 基因型与阿托伐他汀诱导的肌肉损伤严重程度增加有关。在具有CYP3A5*3变异等位基因的个体中,肌酸激酶水平高于野生型个体(P=0.01)[45]。CYP3A基因型与阿托伐他汀诱导肌肉损伤的严重程度增加有关。服用阿托伐他汀的CYP3A5*3/*3基因型患者出现肌痛的可能性更大且更严重[45]。因此,携带CYP3A5*3变异等位基因的个体应减少药物剂量以减轻肌肉损伤程度。发生他汀类相关横纹肌溶解不良反应约60%与药物相互作用有关[46]。临床上使用CYP3A4酶的抑制剂或诱导剂会影响阿托伐他汀的血药浓度,导致发生不良反应的风险增加。
-
他汀类药物的另一种常见不良反应是肝毒性。肝毒性的发生率以血中转氨酶浓度升高为特征,远低于SAMS。ABCB1 rs2032582可预测日本人群患阿托伐他汀诱导的肝损伤(AILI)的风险。ABCB1中的一个SNP位点(rs2032582:2677G>T/A)与AILI显著相关(P=0.00068,OR=2.59,95%CI为1.49~4.50),提示G等位基因可能是AILI的危险因素。利用稳定表达ABCB1 rs2032582编码的ABCB1蛋白的Flp-In-293细胞,研究阿托伐他汀的细胞毒性,明确ABCB1 rs2032582 G等位基因在体内和体外是一个重要的AILI危险因素[47]。ABCB1 rs2032582可能是导致AILI的危险因素,携带该突变基因的个体应减少用药剂量。
-
阿托伐他汀的有关药物代谢酶、转运体、药物作用靶点以及脂质代谢相关基因多态性对阿托伐他汀药动学及药效学的影响,国内外已有很多研究,但仍不完善且许多研究结论不一致,可能是选取人群种族和样本数量的差异造成。目前缺乏多基因相互作用对他汀类药物治疗的影响研究,未来还需参考更多基因多态性以指导临床他汀类药物个体化合理用药。
Research progress of atorvastatin gene polymorphism
doi: 10.12206/j.issn.2097-2024.202110042
- Received Date: 2021-10-15
- Rev Recd Date: 2022-01-21
- Available Online: 2022-09-29
- Publish Date: 2022-09-25
-
Key words:
- atorvastatin /
- blood lipids /
- drug metabolizing enzymes /
- drug transporters /
- gene polymorphisms
Abstract: Atorvastatin is a blood lipid-lowering drug widely used clinically. Long-term use can prevent and reduce the occurrence of atherosclerotic cardiovascular disease (ASCVD). However, the efficacy of atorvastatin has significant inter-individual differences. Some individuals failed to achieve the expected lipid-lowering target value or had serious adverse reactions, which were related to the genetic diversity between individuals. Genetic variation can lead to differences in drug configuration, clinical efficacy and adverse reactions. The drug metabolism enzymes, drug transporters, drug targets and genetic polymorphisms related to lipid metabolism were reviewed in this paper that affect the drug response of atorvastatin, and from the gene level to explore the reasons for the differences in the pharmacokinetics, pharmacodynamics and susceptibility to adverse reactions of different individuals using atorvastatin.
| Citation: | HUANG Qin, SUONAN Gele, LI Wenbin, SUN Yuemei, WANG Rong. Research progress of atorvastatin gene polymorphism[J]. Journal of Pharmaceutical Practice and Service, 2022, 40(5): 416-421, 426. doi: 10.12206/j.issn.2097-2024.202110042 |






 DownLoad:
DownLoad: