-
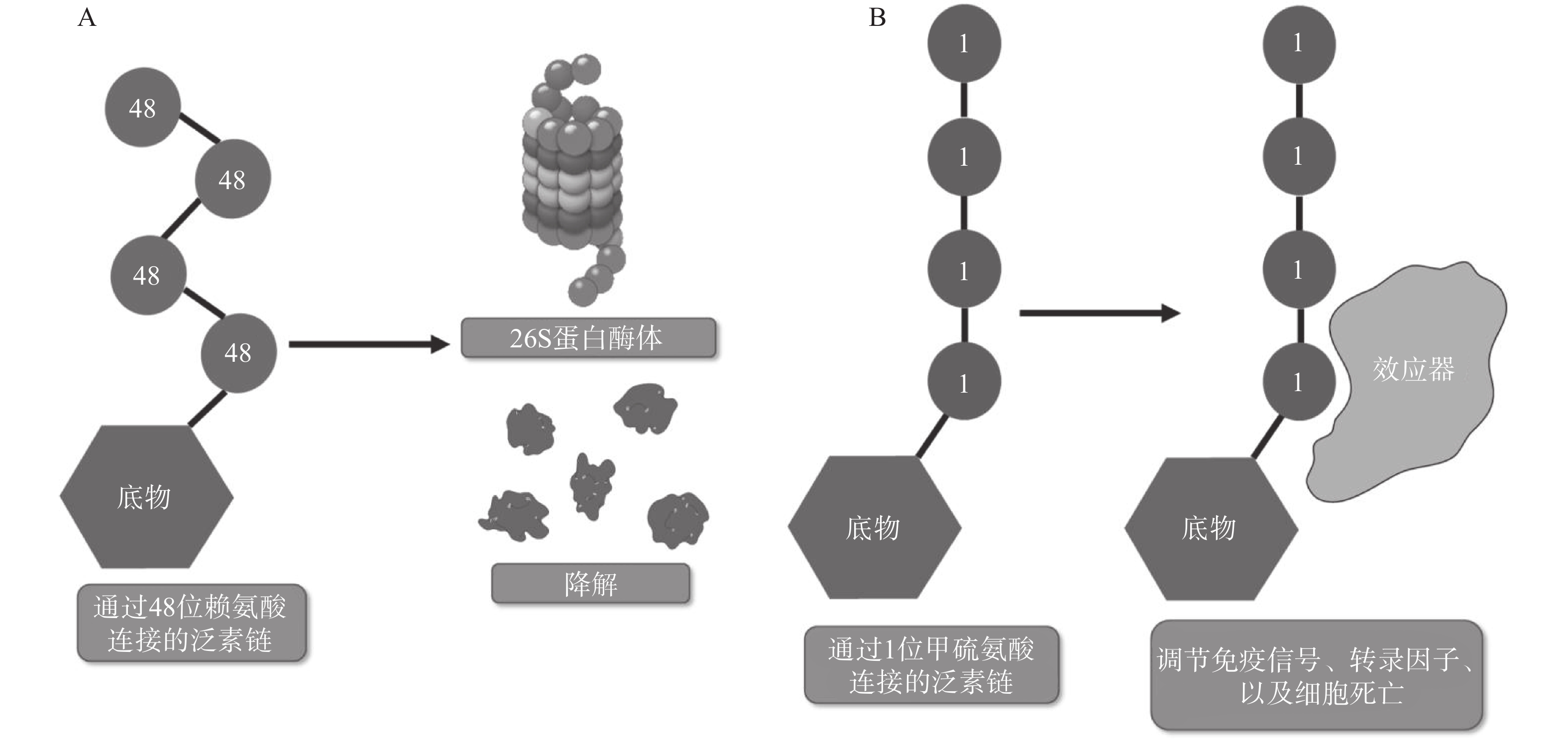
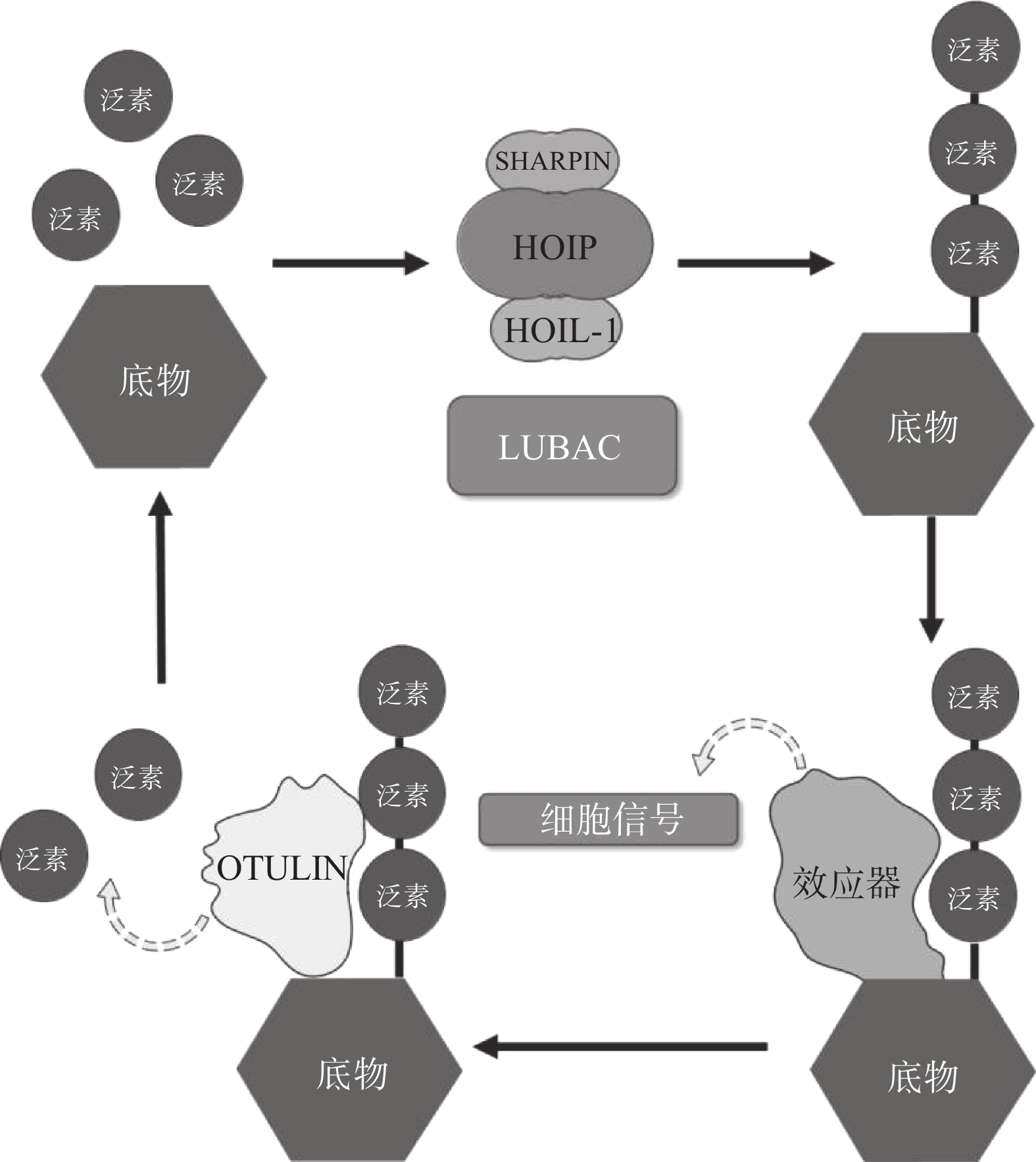
翻译后修饰(PTM)是在蛋白质的氨基酸侧链上共价结合化学小分子基团的过程,能够显著改变蛋白质的理化性质、构象,从而调节、丰富蛋白质的功能。从细胞水平而言,信号蛋白快速而特异性的翻译后修饰,能实现信号的准确传递,引发转录与翻译水平的适应性改变,使细胞及时响应外部环境变化信号,实现自身调节。泛素化修饰是一种重要的翻译后修饰,其中的关键分子——泛素,是一种由76个氨基酸构成的在进化上高度保守的蛋白质,相对分子质量8.5kDa[1]。泛素分子与底物共价结合的过程即泛素化修饰,其能调控基因转录、DNA损伤修复及信号转导等过程[2]。泛素系统的主要功能有两个:一是通过靶向蛋白酶体降解底物或溶酶体降解底物,实现对错误蛋白的调控;二是通过调节蛋白-蛋白相互作用、定位及功能,以实现精细调控细胞信号网络[3]。这种功能上的多样性与泛素化修饰过程中所能募集的不同类型泛素信号有关,根据泛素连接数目的不同,可分为单泛素化修饰和多聚泛素化修饰。在多聚泛素化修饰中,泛素链主要有8种连接模式,其中7种为泛素链内赖氨酸(Lys)与泛素分子C末端甘氨酸(Gly)相连的模式,赖氨酸连接位点包括Lys6、Lys11、Lys27、Lys29、Lys33、Lys48和Lys63,目前研究较多的是Lys48和Lys63位的多聚泛素化修饰。Lys48位的多聚泛素化修饰,其功能是作为蛋白酶体降解的信号,而Lys63位的多聚泛素化修饰主要调控非降解功能,例如参与DNA损伤修复、机体炎症、信号转导等功能[4-6]。其他赖氨酸位点的多聚泛素化修饰功能目前研究较少,但他们在诸如细胞周期调节等过程中,同样是非常重要的胞内信号[7]。剩下的一种连接模式是由一个泛素分子C末端甘氨酸的羧基基团,与另一泛素分子N末端甲硫氨酸(Met1)的氨基基团以肽键相连的模式,形成的是线性泛素链(Linear ubiquitin/Met1-linked ubiquitin chains/Met1-Ub)[8]。近年来的研究表明,线性泛素链在免疫信号调节、NF-κB转录因子激活及细胞凋亡等过程中能发挥强有力的调控作用。更有意义的是,多项研究表明线性泛素链信号的异常调节与免疫失调、肿瘤等人类疾病密切相关[8](图1)。本文将介绍线性泛素连接酶复合体(LUBAC)、去线性泛素化酶(OTULIN)及其与肿瘤的关系。
-
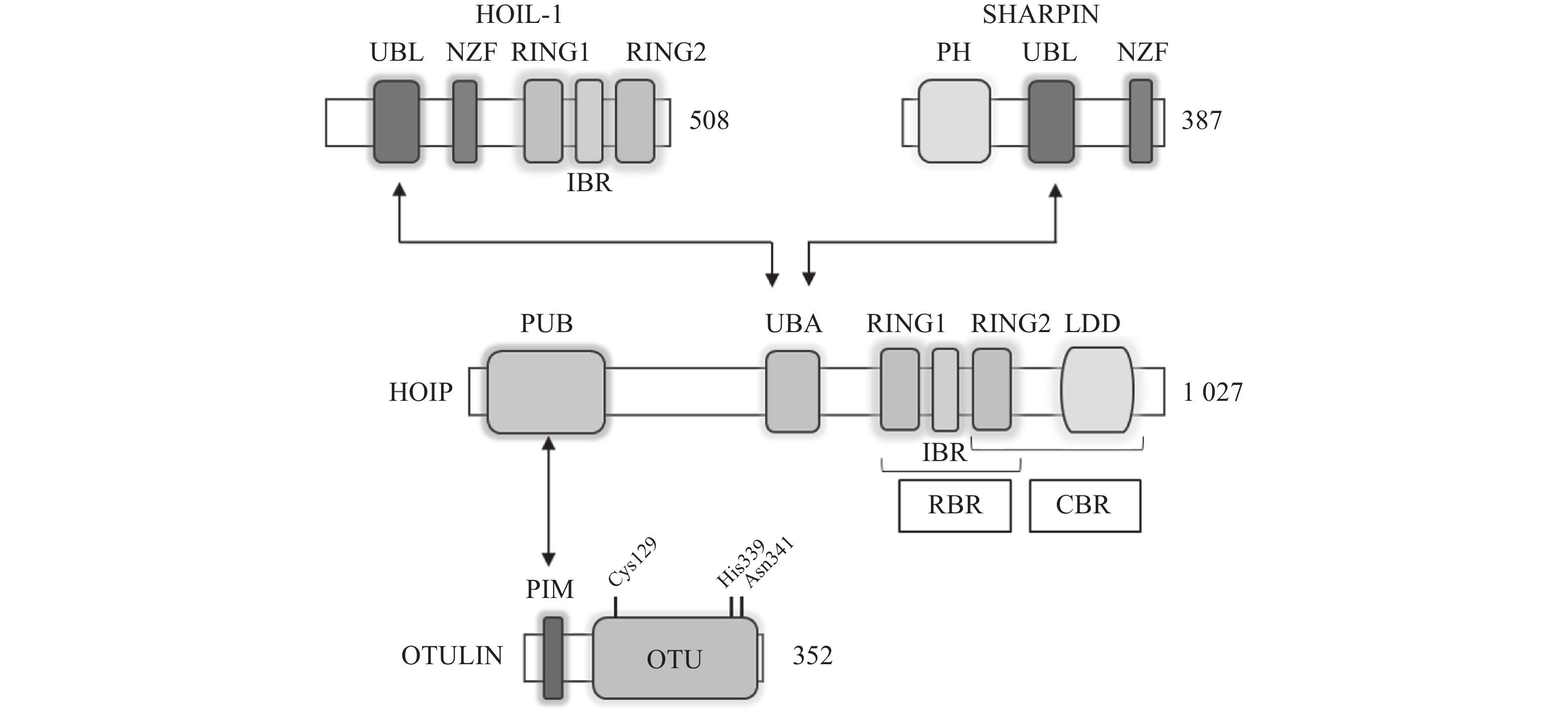
LUBAC是脊椎动物中唯一发现的线性泛素连接酶,该复合体分子量约为600KDa,由三个蛋白构成:HOIP(HOIL-1-interacting protein,RNF31)、HOIL-1(Heme-Oxidised IRP2 Ub Ligase-1,RBCK1)和SHARPIN(SHANK-Associated RH Domain Interacting Protein)(图2)[9]。LUBAC通过这3部分结构的多种分子内相互作用维持稳定,失去任何部分都会影响复合物的稳定性并影响其细胞内活性[8, 10-11]。HOIP与HOIL-1同属RBR(Really Interesting New Gene (RING)-in-between-RING)家族泛素连接酶,两者协同调节LUBAC的活性,尽管两者都有E3泛素连接酶活性,但只有HOIP是主要的E3泛素连接酶[12-13]。然而,HOIP作为单体存在时,会通过RBR、PUB结构域间复杂的分子内相互作用被自抑制[10-11, 14]。HOIL-1和SHARPIN在LUBAC中起到辅助因子的功能,对HOIP的活化至关重要,HOIL-1、SHARPIN的类泛素结构域(UBL),与HOIP的泛素连接结构域(UBA)结合,可以解除HOIP的自抑制,促进线性泛素链的聚集。HOIP的RBR结构域是LUBAC复合物的催化中心[15-16],能通过特殊的RING-HECT杂化反应生成多聚泛素链[17-18]。在线性泛素化的过程中,HOIP的RING1结构域会连接一个带有泛素的E2,这一泛素供体通过瞬间形成硫酯键,被转移到HOIP的RING2结构域中半胱氨酸(CYS)885位点。最后,泛素供体通过HOIP的C端线性泛素链决定域(LDD)与泛素受体结合,特异性地形成线性泛素链[17-19]。
慢性炎症、NF-κB信号通路的激活与癌症的发生、发展、耐药息息相关[20],YANG等证实了LUBAC活性的增强及表达上调,能通过影响NF-κB信号通路,促进活化B细胞样弥漫性大B细胞淋巴瘤(ABC-DLBCL)的发生、发展[20]。罕见的HOIP功能变体在ABC-DLBCL患者中(突变率7.8%)较健康人群(突变率1.0%)增加了近8倍,这种功能变体通过调节HOIP的α螺旋,增加了HOIP与HOIL-1的相互作用,因而提升了LUBAC的活性并激活NF-κB信号通路[20]。同时,在B细胞受体途径(BCR)中,LUBAC与CARD11/MALT1/BCL10复合物的结合,对ABC-DLBCL的生存有重要作用[20]。此外,JO等证实,HOIP的表达在ABC-DLBCL细胞中有整体提升,其能够阻止细胞凋亡、增强NF-κB信号通路并导致AID介导的突变,以此促进淋巴瘤的生成[21]。LUBAC表达及活性的上调增强了BCR介导的NF-κB活性,并保护其免受基因毒性应激诱导的细胞凋亡,以此促进突变体细胞的聚集及淋巴瘤的生成[20-22]。SONG等证实LUBAC在ε蛋白的作用下,能使NF-κB重要调节因子NEMO线性泛素化,可促进NF-κB的持续激活,从而导致雌激素阴性乳腺癌的发生[23]。以上研究表明LUBAC介导的NF-κB的活化可以成为多种肿瘤发生的一种途径。JO等还利用高通量筛选,获得能够特异性抑制LUBAC活性的天然物质——硫藤黄素,在小鼠移植瘤模型中能抑制肿瘤的生长[21]。
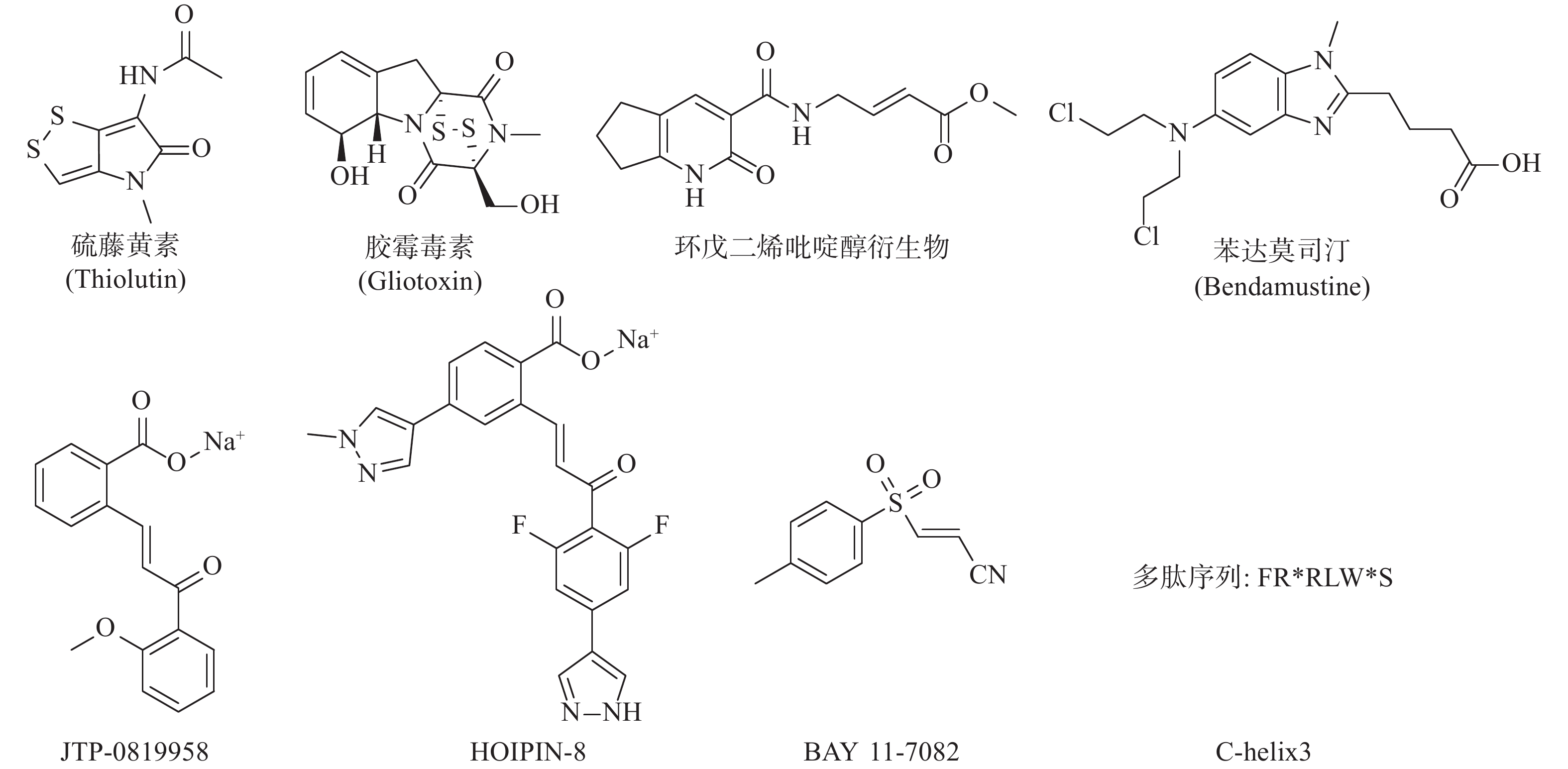
LUBAC介导的NF-κB的激活是人类与小鼠肿瘤耐药的关键因素。RUIZ等人证明了人类与小鼠的肺鳞癌(LSCC)细胞中均存在LUBAC表达水平的上调,这导致了肿瘤中线性泛素链的堆积以及NF-κB活性的增强,从而导致对顺铂耐药的产生[24]。利用LUBAC的小分子抑制剂胶霉毒素,通过抑制LUBAC的活性,降低NF-κB的活性,能够使LSCC细胞重新对顺铂的治疗变得敏感,半剂量的顺铂与胶霉毒素的联合用药,显著抑制了肿瘤在小鼠体内生长,荷瘤小鼠的生存期明显延长,且未显示出毒副作用[25]。同样,HOIP也被证实能够在许多肿瘤株中诱导对顺铂耐药的产生。MACKAY等发现低表达HOIP的细胞会显示出对顺铂的高度敏感性,这主要依赖于ATM介导的DNA损伤检查点的激活,所导致的半胱氨酸蛋白酶8与半胱氨酸蛋白酶3介导的细胞凋亡的增加。HOIP表达量的降低能使对顺铂耐药的卵巢癌细胞,重新恢复对顺铂的敏感性[26]。以上研究表明抑制LUBAC的活性在特定的肿瘤中,可能能够直接或间接地成为有效治疗手段。基于LUBAC在肿瘤发生、发展、耐药过程中的重要作用,研究人员开始了针对LUBAC抑制剂的探索(图3)。除了JO等人报道的硫藤黄素,以及RUIZ等人报道的胶霉毒素以外,JOHANSSON等人利用基于片段的共价配体筛选技术,获得了靶向LUBAC催化亚基HOIP活性位点半胱氨酸的抑制剂,并进一步通过基于细胞的分析和化学蛋白质组学技术,证明以环戊二烯吡啶醇衍生物为代表的一类化合物能有效地渗透到哺乳动物细胞中,标记并抑制HOIP和NF-κB的激活,这些化合物有潜力作为先导化合物,开发选择性探针,以研究LUBAC生物学功能[27]。Virginia等人报告了一种基于MALDI-TOF质谱的无标记高通量筛选方法,能检测抑制剂对泛素E2偶联酶和E3连接酶的抑制强度和特异性,并发现苯达莫司汀是潜在的LUBAC抑制剂[28]。KATSUYA等建立了由LUBAC介导的线性泛素化的细胞非依赖实验,及以NF-κB荧光素酶报告基因为基础的基于细胞的LUBAC实验,两者构成了高通量筛选平台,以此发现LUBAC抑制剂。筛选所得的化合物JTP-0819958能选择性作用于HOIP催化结构域的赖氨酸残基,实现对LUBAC活性的抑制,并在细胞水平降低LUBAC介导的线性泛素链的生成与炎性细胞因子所导致的NF-κB的激活[29]。KATSUYA等以JTP-0819958为先导化合物,测得其LUBAC抑制活性IC50值为2.8 μmol/L,构效关系研究发现其中的α、β-不饱和酮结构是活性必须基团,能通过迈克尔加成反应可逆抑制LUBAC的活性,并由此设计合成了HOIPIN-2~8,最终发现HOIPIN-8有最强的LUBAC(IC50=11 nmol/L)、NF-κB通路的抑制活性,其可能的作用位点同样是HOIP催化结构域的赖氨酸残基,HOIPIN-8是已报道的最有效的LUBAC抑制剂[30]。STRICKSON等人发现NF-κB抑制剂BAY 11-7082能通过使LUBAC失活,发挥抗炎作用、诱导B细胞淋巴瘤和白血病T细胞死亡,并阻止蛋白质向DNA损伤位点募集[31]。AGUILAR-ALONSO等开发了基于HOIP的固定α螺旋的多肽C-helix3,其能够通过与HOIP结合,阻断HOIL-1与HOIP的相互作用,抑制LUBAC活性及功能,并实现降低细胞存活率、减少NF-κB信号通路的激活及其相关基因的产生[32]。虽然目前针对LUBAC抑制剂的研究尚处于初级阶段,但可以预见的是,通过抑制LUBAC活性,影响机体免疫调节,会成为当前及今后肿瘤治疗研究的新方向。
-
OTULIN,是目前唯一能够水解线性泛素链的哺乳动物去泛素化酶,也被称为Fam105b或Gumby,由352个氨基酸构成,其拥有一段含Cys129/His339/Asn341三分子催化中心的高度保守的OTU结构域[33-34],该结构域以底物辅助催化机制发挥功能,即OTULIN仅在与线性泛素链作用时被激活[34]。OTULIN的C端是PDZ(PSD95–Dlg1–ZO-1)结构域结合基序,N端是PUB相互作用基序(PIM),可与HOIP的PUB结构域发生相互作用,发挥去线性泛素化酶的功能(图2)[35- 36]。OTULIN能在不影响其他种类泛素链的情况下,专一性地水解线性泛素链,即使在高浓度OTULIN情况下,也不会作用于与线性泛素链结构高度相似的Lys63连接模式的泛素链,其与线性泛素链的结合常数KD为20 nmol/L,与Lys63连接的泛素链KD为12 μmol/L,两者有超过100倍的结合活性差[34]。OTULIN同样广泛参与机体免疫、信号转导等多个过程,OTULIN的存在可以防止LUBAC复合体的自身泛素化,以及线性泛素链在细胞内的堆积[37-39]。此外OTULIN在TNFα与NOD2响应的刺激下,可以限制LUBAC的活性以及NF-κB通路的激活[39]。OTULIN的N端可通过与蓬乱蛋白2(DVL2)相互作用,与LUBAC共同调节Wnt信号通路[40](图4)。
DAMGAARD等揭示了OTULIN在肝脏稳态与病理状态中的重要作用,作者构建了肝实质细胞内特异性敲除OTULIN的(OtulinΔHEP)小鼠,该模型会自发产生诸如肝炎、肝细胞凋亡、代偿性的肝细胞增生等症状,并导致脂肪性肝炎、肝纤维化以及肝细胞癌(HCC)。Fas相关死亡域(FADD)基因的消除能挽救这一过程,敲入未激活的受体相互作用蛋白激酶1(RIPK1)同样能防止小鼠患上肝脏疾病,这也进一步证明了缺失OTULIN的肝细胞的凋亡,是引发肝脏疾病的关键,OTULIN在肝脏中能预防慢性肝炎及肝细胞癌,起到关键的保护作用[41]。同样,HOSTE等证实了OTULIN在维持皮肤稳态、防止疣状癌的发生中也有重要作用。作者构建了角质细胞特异性缺失OTULIN的小鼠(ΔKerOTULIN),该模型会自发产生炎症性皮肤损伤并发展成疣状癌。在缺失OTULIN的角质细胞中会显示1型干扰素与白细胞介素-1β的响应信号,从基因层面或药物层面抑制这些细胞因子可以一定程度抑制皮肤炎症。此外,表达会引发人类OTULIN相关自身炎症综合征(ORAS)的OTULIN突变基因,会引发相类似的皮肤炎症表型,以此证明OTULIN在抑制皮肤炎症、维持免疫稳态、避免皮肤肿瘤发生中的重要价值[42]。
不同于OTULIN在肝脏、皮肤中的保护作用,WANG等证实OTULIN在一定条件下参与肿瘤耐药的形成。三阴性乳腺癌细胞中Wnt/β-catenin信号通路被异常激活,其在癌症的发展、转移、耐药中发挥着重要作用。在化疗过程中,基因毒性的治疗会导致OTULIN56位酪氨酸的磷酸化,使OTULIN从与HOIP结合中被释放,并增强OTULIN与β-catenin的相互作用,Wnt/β-catenin信号通路以一种OTULIN依赖的方式被进一步活化,诱导耐药的产生。此外,作者还证明了抑制Wnt/β-catenin信号通路或OTULIN能恢复三阴性乳腺癌移植瘤模型对化疗药物的敏感性并减少肿瘤的转移[43]。同时,体内OTULIN水平较高的乳腺癌患者,表现出较低的总体生存率与无疾病生存期。由于Wnt/β-catenin信号通路在正常组织中广泛存在,通过抑制OTULIN以克服肿瘤耐药与转移,是治疗三阴性乳腺癌的一种新选择。
-
线性泛素化修饰作为一种特殊的翻译后修饰,由于其在细胞死亡、免疫信号调节及其他细胞重要功能中所发挥的作用,成为了近年来的研究热点。LUBAC与OTULIN在细胞内可实现对线性泛素链信号的精细调节,而其异常表达则会导致ORAS、肿瘤等严重疾病的发生。目前已能实现利用小分子化合物或多肽对LUBAC的活性进行调节,但其具体作用机制仍有待深入阐明。以HOIPIN-8为代表的高活性LUBAC抑制剂(IC50=11 nmol/L)在体外展现出了较优的抑制效果,然而,需开展更为广泛的体内药效与安全性实验,以验证LUBAC抑制剂的优势,以及LUBAC作为肿瘤、炎症等疾病治疗靶点的潜在价值。对于OTULIN而言,由于其在不同组织中广泛的生理调控功能,未来针对OTULIN的研究重点,仍将停留在对其生理作用与机制的阐明。同时,目前尚无OTULIN小分子配体或抑制剂的报道,因此,开发精确调节OTULIN活性的分子,实现免疫调节或疾病治疗,也是未来极具价值的研究方向。
Research Progress on Linear Ubiquitin Chain Assembly Complex and OTU Deubiquitinase With Linear Linkage Specificity in Tumor
doi: 10.12206/j.issn.2097-2024.202307045
- Received Date: 2023-07-21
- Rev Recd Date: 2023-08-15
- Available Online: 2023-09-25
- Publish Date: 2023-09-25
-
Key words:
- Linear ubiquitination /
- Linear ubiquitin chain assembly complex /
- OTU deubiquitinase with linear linkage specificity /
- Tumor
Abstract: Linear ubiquitination is an important post-translational modification that has been discovered in recent years. The linear ubiquitin chain is formed by the linkage of glycine residue of one ubiquitin protein to the methionine residue of another ubiquitin. This process is regulated by the linear ubiquitin chain assembly complex (LUBAC) and the OTU deubiquitinase with linear linkage specificity (OTULIN). Linear ubiquitination is involved in various biological processes, including immune response, inflammation, and cell apoptosis. Recent studies have shown that linear ubiquitination is closely related to the occurrence, development, and drug resistance of tumors by affecting signaling pathways such as NF-κB and Wnt/β-catenin. The research progress on the function of LUBAC and OTULIN in tumors was reviewed in this paper.
| Citation: | FANG Yuxin, LI Yu, LIU Baoshu, DONG Guoqiang. Research Progress on Linear Ubiquitin Chain Assembly Complex and OTU Deubiquitinase With Linear Linkage Specificity in Tumor[J]. Journal of Pharmaceutical Practice and Service, 2023, 41(9): 534-539. doi: 10.12206/j.issn.2097-2024.202307045 |






 DownLoad:
DownLoad: