-
细菌耐药性已成为全球公共卫生威胁,其中耐碳青霉烯类肠杆菌目(CRE)细菌的感染是全球抗感染领域最引人注目的问题之一,原因是临床仅以碳青霉烯类抗菌药物已无法有效治疗此类细菌的感染[1],尤其是新德里金属β-内酰胺酶(NDM)的出现,给防控耐药菌株的传播敲响了警钟,到目前为止,临床可用于治疗产NDM型碳青霉烯酶菌株的药物仍寥寥无几[2-3]。2021年,我国肺炎克雷伯菌(CR-Kpn)对亚胺培南和美罗培南的耐药率分别为20.8%和21.9%,几乎是2005年的7倍(3.0% 和2.9%)。大肠埃希菌(CR-Eco)对亚胺培南和美罗培南的耐药率分别为1.8%和2.0%[4]。此外,研究表明,新冠肺炎可能会加速CRE通过病毒促进细菌附着和呼吸道定植,从而导致CRE在世界各地的传播率有所上升[5-6]。肠杆菌目不同属细菌的耐药性可能由多种机制单独或协同介导,不同菌株主要的耐药机制也不尽相同,对于特定菌株来讲,各个机制之间的协同作用也会大大提高其对碳青霉烯类抗生素的耐受性[7-8]。因此,为应对CRE带来的重大挑战,实验室需做好抗微生物药物敏感性试验,开展针对碳青霉烯类耐药革兰阴性杆菌的酶型检测,为临床的抗感染治疗提供尽其所能的协助。本研究旨在通过评价上海交通大学医学院附属仁济医院浦南分院(本院)CRE对常见临床抗菌药物体外敏感性药敏试验的结果,了解CRE最常见的基因型,以及比较碳青霉烯酶耐药表型和基因型两种不同检测方法,以期为CRE的临床治疗和医院感染控制提供流行病学依据。
-
菌株来源:收集本院2022年1月至12月患者临床标本分离的CR-Kpn及CR-Eco非重复分离株,共400株。
-
使用珠海迪尔 Smart MS质谱仪对菌株进行鉴定。体外药敏试验使用96孔微量肉汤稀释法(仪器:Nephelometer比浊仪、Sensititre AIM自动加样系统、肉汤阅读仪)。
-
美罗培南、环丙沙星、多黏菌素B、阿米卡星、替加环素均用无菌水溶解及稀释;头孢他啶用磷酸缓冲液(pH=6.0)溶解,无菌水稀释;头孢吡肟均用磷酸缓冲液(pH=6.0)溶解及稀释;氨曲南用饱和碳酸氢钠溶解,无菌水稀释。
-
将过夜纯分培养的受试菌用直接菌落悬浮法调制成0.5麦氏浊度管比浊,行100倍稀释,最终接种菌量为105 CFU/ml。
-
肠杆菌目:空气状态下,(35±2)℃培养16~20 h。
-
最低抑菌浓度(MIC)结果根据美国临床和实验室标准化协会2022年M100第32版文件颁布的折点进行判读[9]。CRE的定义是根据美国疾病预防控制中心2015年颁布的文件[10]。
-
CR-Eco ATCC25922、CR-Kpn ATCC700603 (上海市临床检验中心)。
-
使用RESIST-5 O.O.K.N.V.(CORIS Bioconcept)胶体金法,对碳青霉烯类药物耐药(美罗培南MIC值≥4)的菌株,进行初筛。
-
采用聚合酶链反应(PCR)技术检测CR-Eco blaKPC、blaNDM;CR-Kpn blaKPC、blaNDM、blaOXA-48 耐药基因,对阳性扩增产物进行测序确认。
-
采用煮沸裂解法制备细菌DNA样本,即从培养基上选取新鲜培养的单个菌落到0.5 ml双蒸水,100℃震荡加热15 min,之后10 000 r/min离心5 min,留上清液作为被检测DNA样本。
-
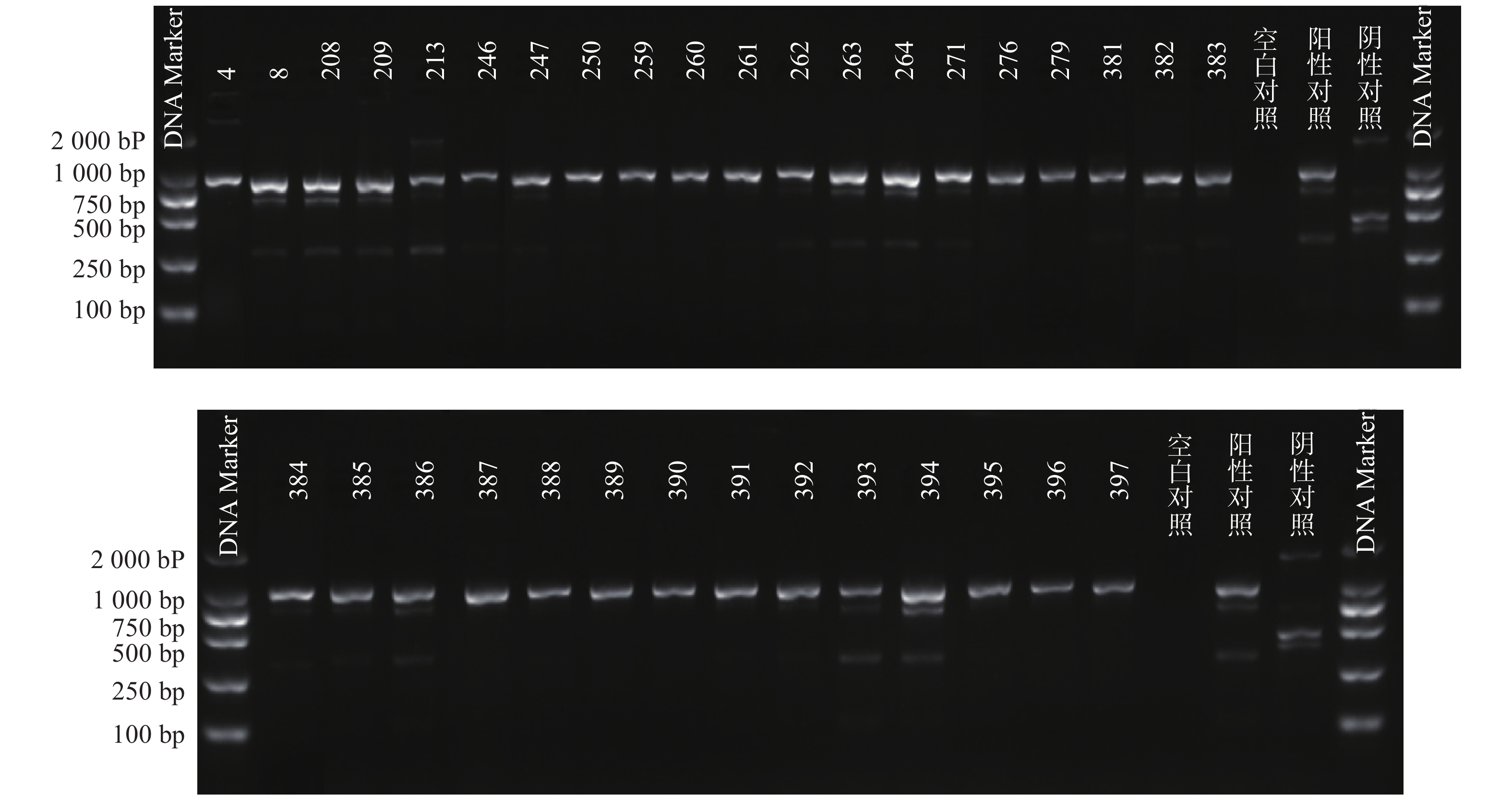
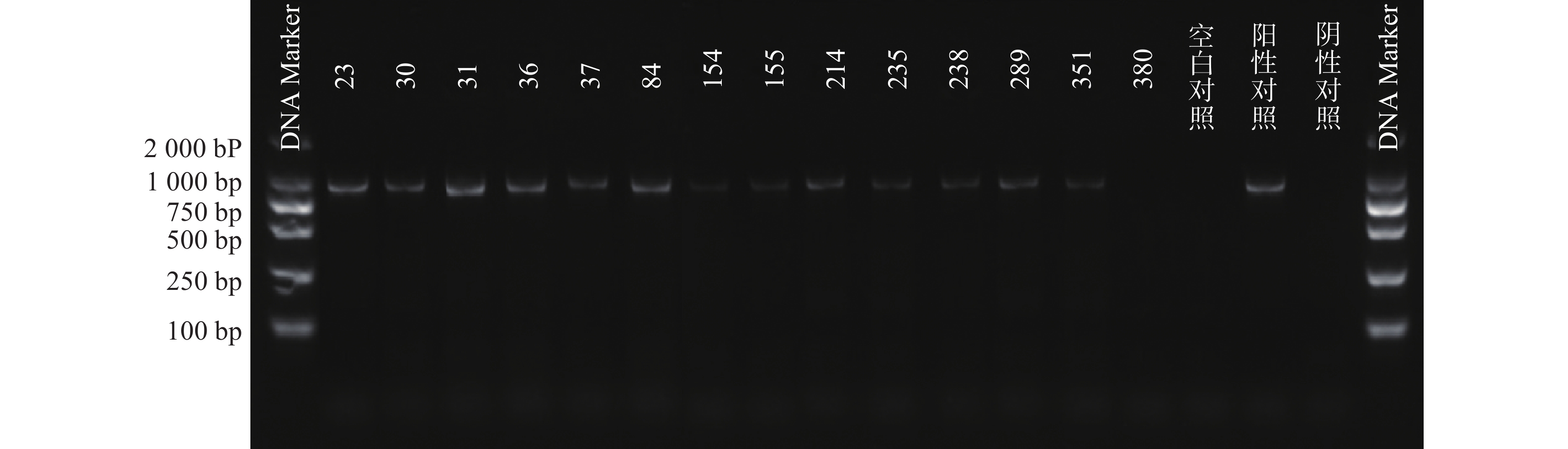
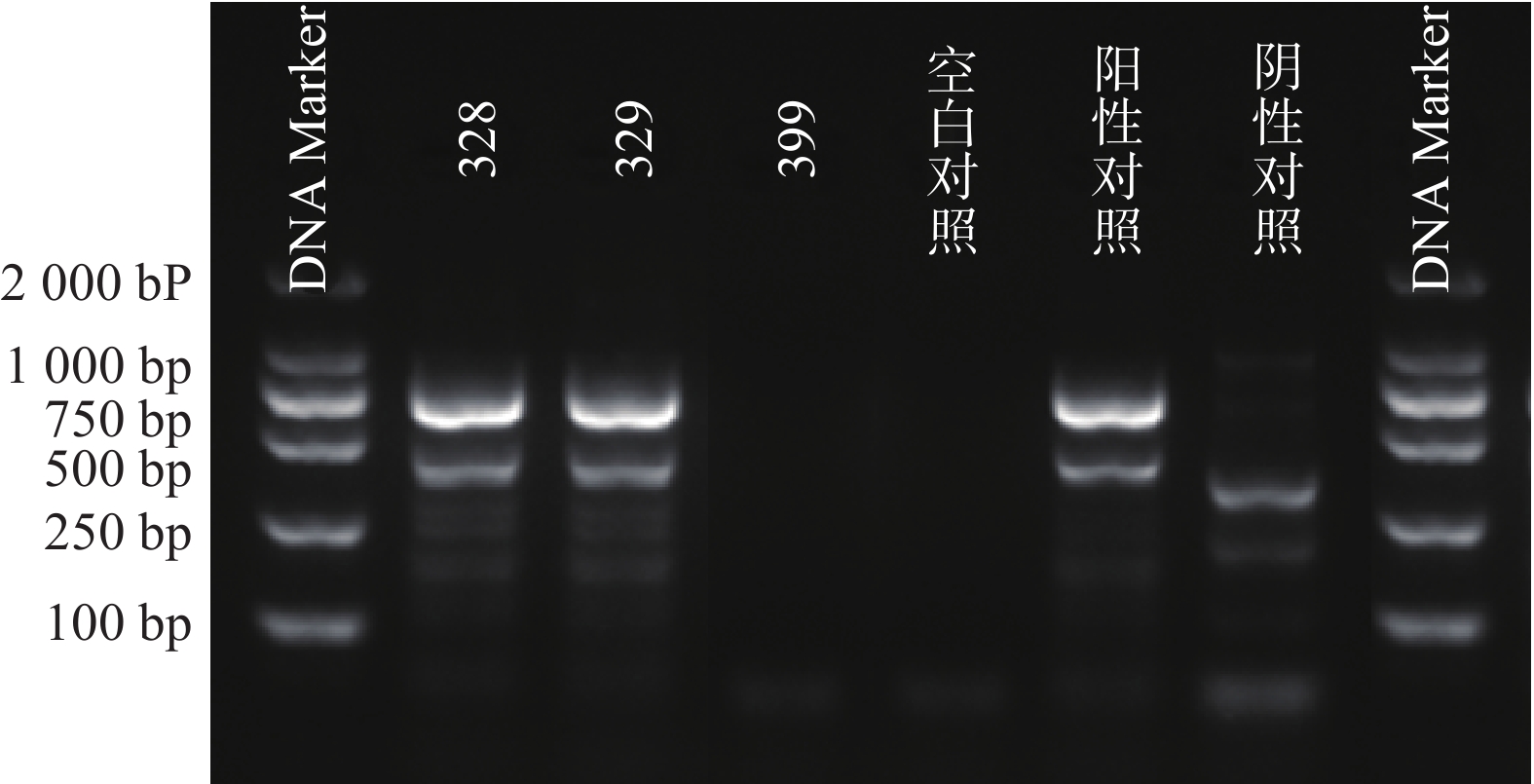
根据参考文献设计blaKPC、blaNDM 、blaOXA-48 3个耐药基因的检测引物(购自上海生工生物工程技术有限公司)。TaKaRa TaqTM HS Perfect Mix、DL2000DNA Marker等PCR试剂(TakaRa Bio)。PCR反应条件为:95℃预热5 min;95℃变性30 s,58℃退火30 s,72℃延伸40 s,共38次循环;72℃反应5 min,然后4℃储存。PCR反应完成后,取5 μl PCR产物与1 μl 6×上样缓冲液混合后加入2.5%琼脂糖凝胶上样孔中进行电泳,设恒压110 V电泳30 min。最后通过凝胶成像仪观察电泳结果。KPC耐药基因的产物长度为882 bp、NDM 耐药基因的产物长度为813 bp、OXA-48耐药基因的产物长度为743 bp。
-
PCR电泳后观察,将符合预期片段长度的产物进行测序验证。
-
采用WHONET 5.6版软件对药敏试验结果进行统计分析,其中计数资料以例数和百分比表示。
-
共分离到CR-Kpn及CR-Eco非重复分离株共400株,其中CR-Kpn 195株(48.75%),CR-Eco 205株(51.25%)。51株CRE中CR-Kpn 41株(80.39%)、CR-Eco 10株(19.61%)。CRE菌株大多来自呼吸道,血液和中段尿标本。其中,CR-Kpn来源的前3位标本分别为:痰标本、血标本、中段尿标本,CR-Eco来源的前3位标本分别为:中段尿标本、血标本、导管,具体见表1。此外,41株CR-Kpn主要分布在ICU 21株(51%)和老年科5株(12%);10株CR-Eco主要分布在普外科6株(60%)。
标本类型 CR-Kpn(n=41) CR-Eco(n=10) 株数 构成比(%) 株数 构成比(%) 痰 17 41.46 0 0.00 血液 11 26.83 3 30.00 中段尿 8 19.51 6 60.00 肺泡灌洗液 2 4.88 0 0.00 导管 0 0.00 1 10.00 脓 1 2.44 0 0.00 胆汁 1 2.44 0 0.00 胸水 1 2.44 0 0.00 -
CR-Kpn对美罗培南耐药率为21.03%、对替加环素、多黏菌素B、阿米卡星耐药率为0.00%、4.10%、11.79%,对头孢吡肟、头孢他啶耐药率分别为48.21%、45.64%,对氨曲南、环丙沙星耐药率分别是50.26%、53.85%。CR-Eco对美罗培南耐药率为4.88%,对替加环素、多黏菌素B、阿米卡星耐药率分别为0.00%、0.49%、0.98%,对头孢吡肟、头孢他啶耐药率分别为30.24%、39.51%,对氨曲南、环丙沙星耐药率分别是41.95%、64.88%,见表2。
抗菌药物 肺炎克雷伯菌菌(n=195) 大肠埃希菌(n=205) 耐药 敏感 耐药 敏感 美罗培南 21.03 78.97 4.88 95.12 头孢吡肟 48.21 43.59 39.51 48.78 头孢他啶 45.64 48.72 30.24 62.44 环丙沙星 53.85 41.54 64.88 22.44 替加环素 0.00 97.44 0.00 100.00 多黏菌素B 4.10 95.90 0.49 99.51 阿米卡星 11.79 88.21 0.98 99.02 氨曲南 50.26 47.69 41.95 48.78 注:替加环素折点参考FDA,多黏菌素折点参考杨启文等[11]专家共识,其余均参考CLSI。 -
对 CRE的体外抗菌活性最高的前2位抗菌药物分别为替加环素(0.00%)、多黏菌素B(7.84%)。此外,对CR-Eco体外抗菌活性较高的还包括阿米卡星(90%)和氨曲南(80%),见表3。
抗菌药物 肺炎克雷伯菌菌(n=41) 大肠埃希菌(n=10) 耐药 敏感 耐药 敏感 美罗培南 100.00 0.00 100.00 0.00 头孢吡肟 100.00 0.00 90.00 0.00 头孢他啶 100.00 0.00 100.00 0.00 环丙沙星 100.00 0.00 100.00 0.00 替加环素 2.56 97.44 0.00 100.00 多黏菌素B 10.26 89.74 0.00 100.00 阿米卡星 51.28 48.72 10.00 90.00 氨曲南 100.00 0.00 10.00 80.00 注:替加环素折点参考FDA,多黏菌素折点参考杨启文等[11]专家共识,其余均参考CLSI。 -
51株CRE中检测到碳青霉烯酶49株(96.08%)。耐药表型检测中产NDM型碳青霉烯酶共13株(25.49%)、产KPC型碳青霉烯酶共34株(66.67%)、产OXA酶共2株(3.92%),见表4。耐药基因检测中以产blaKPC为主共34株(66.67%)、以产blaNDM共13株(25.49%)、以产blaOXA-48为主共2株(3.92%),见表5,图1、图2和图3。
耐药表型 CR-Kpn(n=41) CR-Eco(n=10) KPC酶 32(78.05) 2(20.00) NDM酶 5(12.20) 8(80.00) OXA酶 2(4.88) 0 阴性结果a 2(4.88) 0 注:a表示未检测到任何目标碳青霉烯酶。 耐药基因 CR-Kpn(n=41) CR-Eco(n=10) blaKPC 32(78.05) 2(20.00) blaNDM 5(12.20) 8(80.00) blaOXA-48 2(4.88) 0 阴性结果a 2(4.88) 0 注:a表示未检测到任何目标碳青霉烯酶。 -
近年来,由于抗菌药物的选择局限性和感染控制措施的实施不足,多重耐药菌感染率不断升高,尤其随着碳青霉烯类抗菌药物的广泛应用,在抗生素选择压力下CRE菌株不断被检出,已引起临床关注[12]。肠杆菌目中有许多种属可检测出CRE,最常见的有CR-Kpn、CR-Eco,其次是阴沟肠杆菌、变形杆菌属和弗劳地柠檬酸杆菌等[13-15]。CRE对碳青霉烯类药物的耐药机制主要以产碳青霉烯酶为主,其次是孔蛋白缺失或改变、外排泵过表达、青霉素结合蛋白改变和生物膜产生。耐药基因的突变、插入和转录修饰也可能影响肠杆菌目细菌对碳青霉烯类的敏感性[16]。本研究结果显示51株CRE包括CR-Kpn 41株(80.39%)和CR-Eco 10株(19.61%)。通过耐药表型筛选并结合基因型检测,本研究发现CRE耐药表型与携带的耐药基因型基本一致,CRE菌株中产blaKPC共34株(66.67%)、产blaNDM共13株(25.49%)、产blaOXA-48共2株(3.92%),blaKPC-2基因是本院主要的碳青霉烯类耐药基因,与PORRECA等[17]报道一致。此外,本研究结果显示,CR-Kpn中有78.05%产blaKPC, 12.20%产blaNDM, 4.88%产blaOXA-48,该结果与 CHINET 2016~2018年对全国24个省市的36家医院收集到935株非重复 CRE 菌株研究[18]结果CR-Kpn中产blaKPC 64.6%,产blaNDM 9.5%,产blaOXA-48 4.88%略有不同,可能与当地流行的 CRE携带的碳青霉烯酶耐药基因存在区域差异性有关。本研究体外抗菌药物敏感性试验结果显示,CRE均呈多重耐药,对头孢菌素类、碳青霉烯类出现较高程度的耐药,多黏菌素B,替加环素和一些氨基糖苷类药物是少数可能对CRE保持活性的药物之一。本研究的结果显示替加环素对所有CRE菌株具有较好的抗菌活性,敏感性均>95%,CR-Kpn对多黏菌素B敏感性为89.74%,略低于2021年CHINET监测数据,而CR-Eco对多黏菌素B敏感性为100%。某些抗菌药物对特定CRE菌株有较好的抗菌活性,如CR-Eco对阿米卡星敏感性为90%。由于碳青霉烯类耐药革兰阴性菌往往对临床常用抗菌药物耐药,其所致感染临床治疗选择药物有限,建议临床根据体外药物敏感性试验结果联合用药,如替加环素联合氨基糖苷类或多黏菌素B,但替加环素和多黏菌素B的临床治疗效果可参考的文献较少,建议临床根据实际治疗效果不断调整治疗方案。
本研究也存在某些不足之处,首先,纳入耐药菌除CR-Kpn、CR-Eco外,其他肠杆菌目细菌均未纳入;其次,CRE耐药基因型可能同时携带一种或多种AmpC酶和超广谱β-内酰胺酶[19],其复杂的基因型及其内在表达机制有待于进一步研究。
综上所述,替加环素、多黏菌素B还保持着较高的抗菌活性,KPC-2是本院CRE的主要酶型。由于作用不同碳青霉烯酶抗菌药物对不同碳青霉烯酶的抑制作用不同,因此,对于临床分离的碳青霉烯类耐药肠杆菌目细菌,实验室应开展碳青霉烯酶表型或基因型的检测并进行临床报告。临床应根据微生物实验室药物敏感性及碳青霉烯酶分析结果结合本地区CRE耐药基因型分布特点,制定治疗策略和预防措施,合理用药。
Analysis of resistance situation and resistance genes of clinical isolates of carbapenem-resistant Klebsiella pneumoniae and Escherichia coli
doi: 10.12206/j.issn.2097-2024.202309059
- Received Date: 2023-09-26
- Rev Recd Date: 2024-06-08
- Available Online: 2024-10-24
- Publish Date: 2024-10-25
-
Key words:
- carbapenem-resistant enterobacteriaceae /
- phenotype of carbapenemase /
- carbapenemase resistance gene /
- resistance
Abstract:
| Citation: | HUANG Yun, ZHANG Zhengyin, JIN Ying, ZHENG YiJing, LI Tiejun, SUN Lili. Analysis of resistance situation and resistance genes of clinical isolates of carbapenem-resistant Klebsiella pneumoniae and Escherichia coli[J]. Journal of Pharmaceutical Practice and Service, 2024, 42(10): 439-444. doi: 10.12206/j.issn.2097-2024.202309059 |






 DownLoad:
DownLoad: