-
荧光共振能量转移(fluorescence resonance energy transfer,FRET)是指供体荧光分子发射光谱与受体分子的吸收光谱有显著的重叠且分子间距小于10 nm时发生的一种非放射性的能量转移[1],导致供体荧光淬灭而受体荧光增强或不变。近些年,FRET技术以其精准高效的特点被广泛应用在分析检测领域,为检测生物分子提供了重要的分析方法。基于FRET技术,实现了活细胞内ATP分子的检测[2],金属离子如汞离子的检测[3-4],许多疾病相关基因[5-6]以及酶活性的检测等[7-8]。
银纳米簇(AgNCs),作为一种新型的低毒性“绿色”荧光标记材料,具有量子产率高、毒性低、生物相容性好等特点[9],使得其被广泛应用在多个研究领域。在银纳米簇的合成过程中,相较于其他的合成模板,DNA更具优势,如DNA具有分子识别的功能(包括对于互补链和小分子的识别),不同序列的DNA模板可调谐不同的发射波长等[10]。
研究发现,G碱基可以增强DNA/银纳米簇(DNA/AgNCs)的荧光强度[11-12],基于此,我们设计了系列非银簇模板部分的互补链,考察G碱基个数对于银簇荧光强度的影响,实验结果显示,暴露的G碱基个数与银簇的荧光强度呈正相关关系。该实验不仅验证了G碱基对于银簇荧光的增强作用,还提示我们在设计含有G-四联体适配体的荧光探针时,可通过改变非银簇部分的互补链长短来调控荧光的淬灭及恢复程度,以获得最佳的检测效果。
-
VS-100C恒温混匀仪(无锡沃信仪器制造有限公司);FL-6500荧光分光光度计(PerkinElmer);ZEN3600粒径电位测定仪(英国马尔文公司);精密电子天平(北京赛多利斯仪器系统有限公司);Vortex-Genie2多功能旋涡混合器(美国Scientific Industries公司);TGL-16C离心机(上海安亭科学仪器厂);实验室pH计 FE20[梅特勒-托利多仪器(上海)有限公司];Tecnai G2 F30高分辨率电子显微镜(荷兰FEI公司)
-
硝酸银、硼氢化钠、盐酸(国药集团化学试剂有限公司);三羟甲基氨基甲烷(Tris,大连美仑生物技术有限公司);氯化镁、氯化钠、氯化钾(上海泰坦科技股份有限公司);试剂均为分析纯。实验用水为屈臣氏蒸馏水。
相关DNA序列由生工生物工程(上海)股份有限公司合成。合成DNA-AgNCs的模板序列及P1A5C5的互补序列如表1、表2所示。
表 1 DNA/AgNCs的模板序列
名称 序列(5′—3′) P1C5 GGAGGTGGTGGGGCCCCCTAATTCCCCC P1AC5 GGAGGTGGTGGGGACCCCCTAATTCCCCC P1A5C5 GGAGGTGGTGGGGAAAAACCCCCTAATTCCCCC P1N GGAGGTGGTGGGGCCCTAACTCCCC P1Y GGAGGTGGTGGGGCCCTTAATCCCC 表 2 P1A5C5的互补序列
名称 序列(5′—3′) 0G CCTCCACCACCCCTTTTT 1G CTCCACCACCCCTTTTT 2G TCCACCACCCCTTTTT 4G TCCACCCCTTTTT 5G CACCCCTTTTT 6G ACCCCTTTTT 7G CCCTTTTT -
加入相应体积的20 mmol/L Tris-HCl(pH=7.4)缓冲溶液将DNA溶解,即制得100μmol/L DNA溶液,将制得的DNA溶液95 ℃加热5 min后,冰水浴冷却10 min。
-
参考文献中的合成方法[13],将一定体积的硝酸银溶液加入到上述DNA溶液中(20 mmol/L Tris-HCl,pH=7.4),充分震荡混匀,25 ℃孵育20 min,静置,将一定体积新配制的硼氢化钠引入到上述反应混合物中,最终使得体系中DNA、硝酸银、硼氢化钠的浓度分别为5、30、30μmol/L(即DNA:Ag+∶NaBH4的摩尔比为1∶6∶6),剧烈震荡混匀,室温下避光反应3 h后,4 ℃避光反应过夜。得到的银纳米簇溶液在4 ℃保存以备用。
-
荧光图谱表征:将2μmol/L的银纳米簇溶液在200~800 nm波长范围内进行荧光光谱预扫描,设置激发和发射狭缝宽度为10 nm,扫描速度为1200 nm/min,电压值为400 V。
高分辨率透射电子显微镜表征:将银纳米簇溶液滴加铜网后观察。
-
在含有100 mmol/L NaCl,2 mmol/L MgCl2,5 mmol/L KCl的20 mmol/L Tris-HCl缓冲溶液中(pH=7.4),加入银簇溶液(2μmol/L),将互补的DNA溶液(表2)以1∶1的摩尔比分别加入到上述体系溶液中,充分震荡混匀,37 ℃孵育20 min。在室温条件下进行荧光光谱测量。激发波长为486 nm,激发和发射狭缝宽度为10 nm,扫描速度为1200 nm/min,电压值为400 V。
-
在进行DNA/AgNCs的合成时,基于胞嘧啶碱基与银离子的作用,一般选择富含胞嘧啶碱基的序列作为合成模板,笔者根据文献报道的模板序列结合模板优化设计[13-16],选择了5种银簇模板并进行了相应的碱基优化设计,如表1所示,结果显示P1A5C5的荧光信号强度较高(如图1),且4 ℃避光保存45 d后,荧光信号强度基本不变,稳定性良好。因此,我们使用该序列合成的DNA/AgNCs进行后续的实验研究。
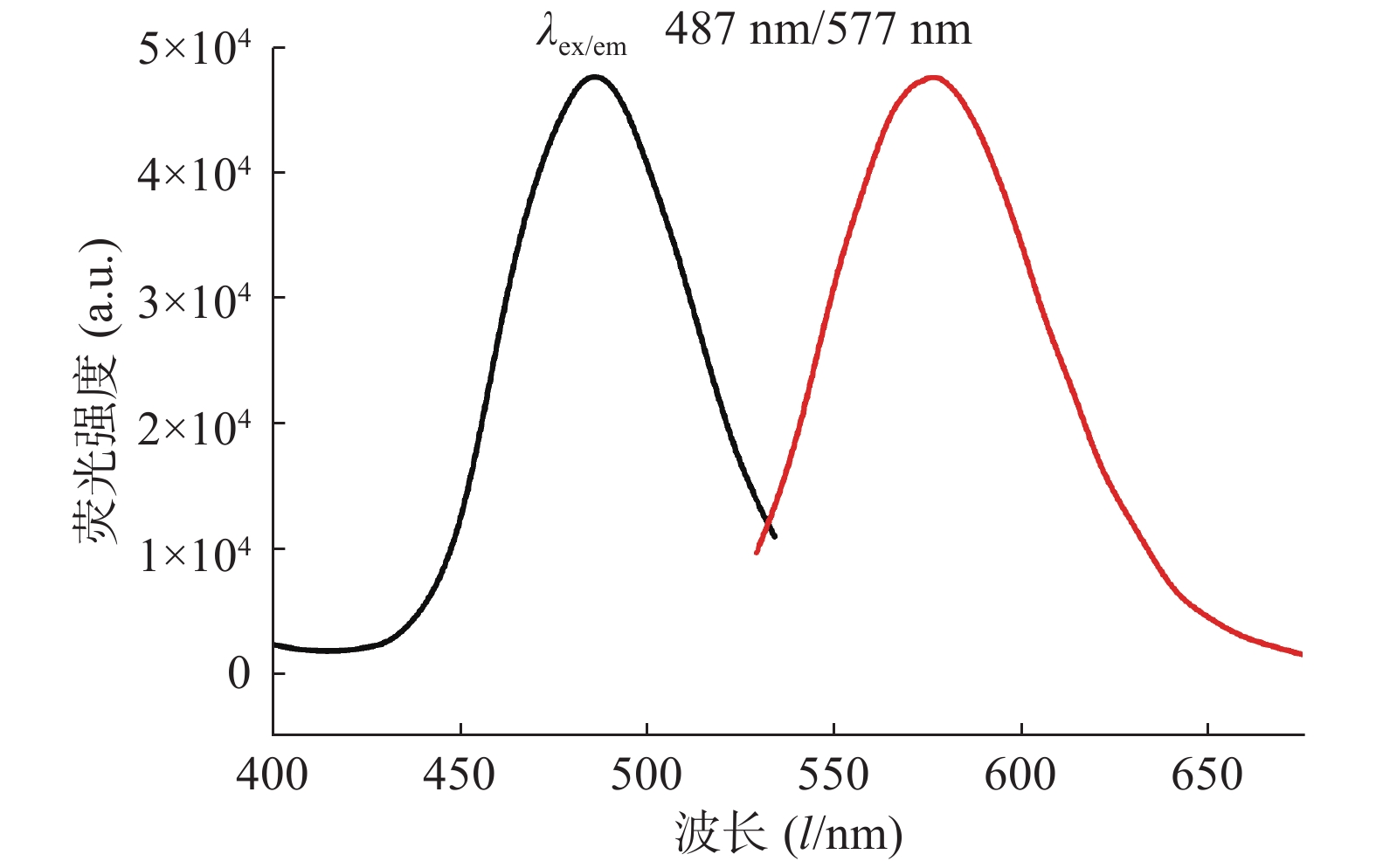
对生成的P1A5C5银纳米簇体系进行荧光激发光谱和发射光谱的表征,结果如图2所示,在487 nm激发条件下,发射波长为577 nm。
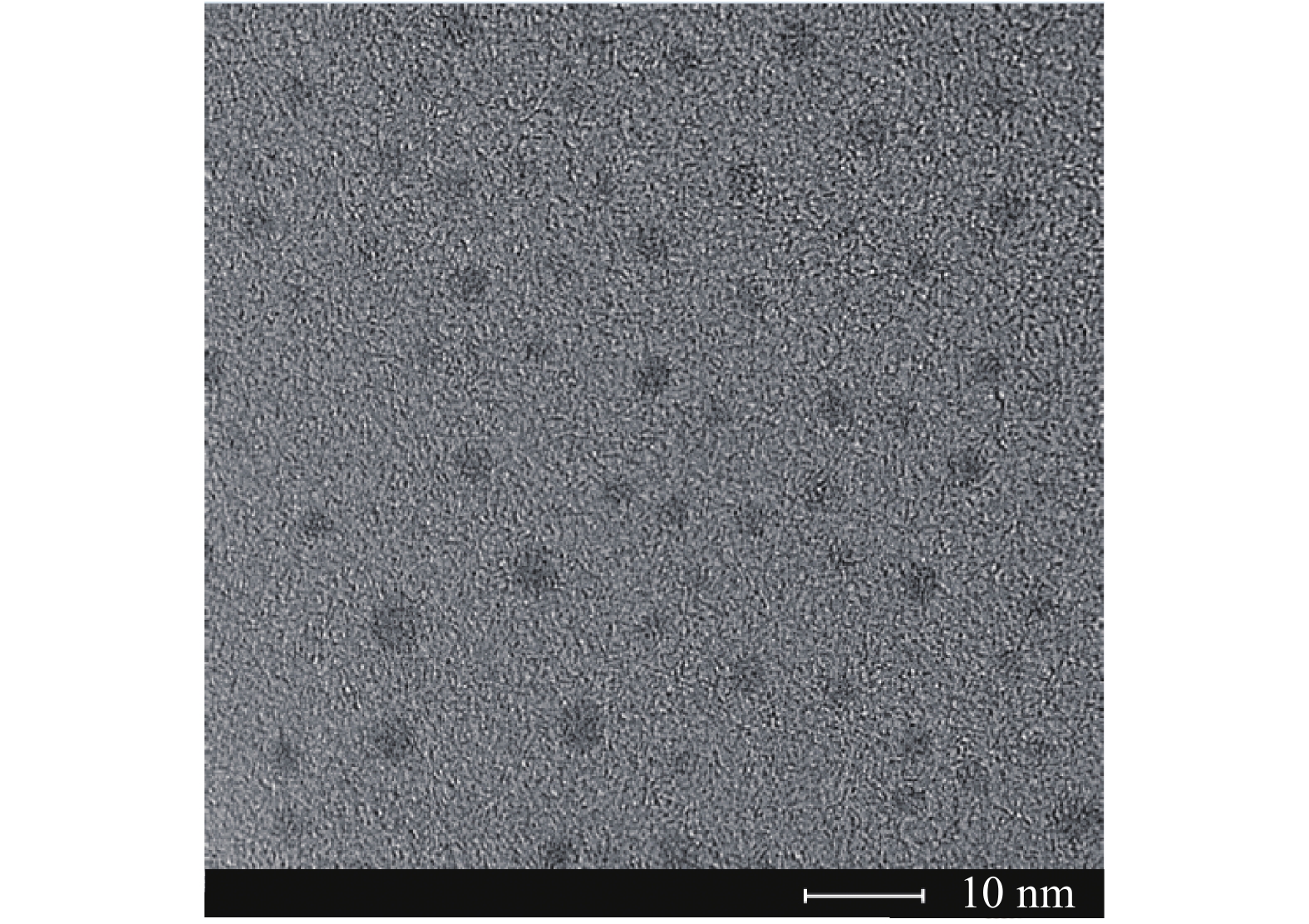
DNA/AgNCs的高分辨透射电子显微镜(HRTEM)图片如图3所示,从图中可以看出,DNA/AgNCs的直径约为2~3 nm,分散性良好。
-
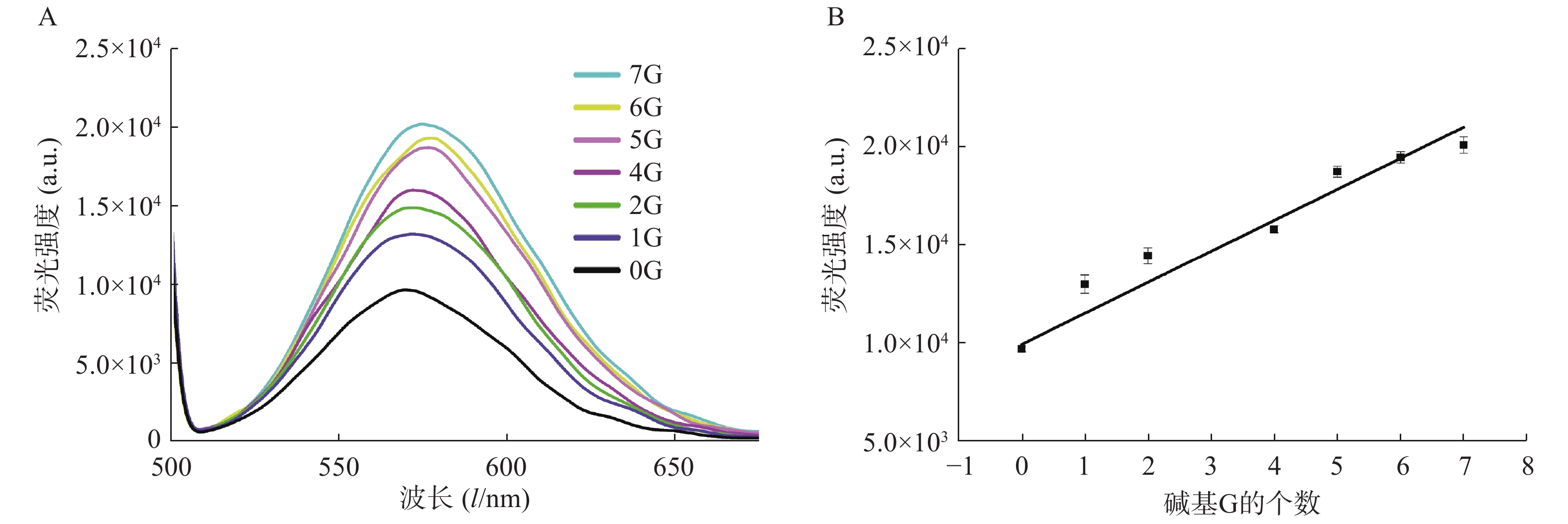
在银簇P1A5C5序列5′-GGAGGTGGTGGGGAAAAACCCCCTAATTCCCCC-3′中,CCCCCTAATTCCCCC为成簇模板序列,下划线部分为岩沙海葵毒素的G-四联体适配体[17]。基于G碱基对银簇的荧光信号强度有增强作用这一现象,我们设计了一系列适配体部分的互补序列(表2),通过C-G碱基互补,适配体部分暴露的G碱基个数不同,导致与银纳米簇作用的碱基G数目不同,进而对银纳米簇的荧光信号强度产生不同的影响。结果显示,随着暴露的G碱基个数的增加,荧光信号强度增加(图4A),对暴露的G碱基个数与荧光信号强度关系进行拟合,得到的线性方程为:Y=1726.1X+8972.5,r=0.9789(图4 B)。该研究证明了G碱基对银簇的荧光具有增强效果,反之,通过C-G碱基互补配对,G碱基与银簇的作用位点被占据,无法与银簇作用,难以达到增强荧光信号强度的作用。因此,随着暴露碱基个数的减少,荧光强度减弱。基于此,可实现与G-四联体适配体有关的荧光开关的设计,进而实现对目标物的检测。
在DNA序列和银纳米簇进行碱基互补配对时,结合互补序列的Tm值,我们研究了互补链间实现碱基互补配对所需的温度和时间。实验结果显示,相较于4 ℃,在37 ℃条件下孵育,互补链间相互作用较强,容易实现碱基互补配对,荧光信号强度变化较为明显;同时,考察了在37 ℃下作用1 h内的荧光变化情况,结果显示,荧光变化强度随时间未发生明显变化,最终选择20 min作为孵育时间。
-
该实验利用含有G四联体适配体的DNA序列合成了荧光银纳米簇,基于G碱基可以增强银纳米簇的荧光这一现象,并结合碱基互补配对的原则,设计了8种G四联体适配体的互补链,在互补链和适配体部分碱基互补配对后,荧光信号强度也产生相应的变化。随着互补链长度的缩短,暴露的G碱基个数增加,荧光信号增强,且暴露的G碱基个数与荧光信号强度拟合得到的线性方程为:Y=1726.1X+8972.5,r=0.9789。该实验不仅证实了G碱基对荧光银纳米簇有荧光增强作用,还提示我们在设计含有G四联体的银纳米簇荧光探针时,可以通过互补链对荧光信号的干扰程度来调控荧光的开闭,这对进一步扩展银纳米簇在荧光分析方法中的应用具有指导意义。
The effect of different guanine base number on fluorescence intensity of DNA/ silver nanoclusters
-
摘要:
目的 通过C-G碱基互补配对的方式,考察不同鸟嘌呤碱基(G)数目对DNA/银纳米簇荧光信号强度的影响,以此来探究新的荧光探针开关构建的方法。 方法 利用核酸碱基互补配对原则,设计了一系列银纳米簇的适配体部分的互补序列,考察了银纳米簇的适配体序列中暴露的G碱基个数对荧光信号的影响。 结果 碱基G可增强银纳米簇的荧光信号强度,且荧光信号强度与G碱基个数呈现正相关关系,拟合线性方程为Y=1726.1X+8972.5,r=0.9789。 结论 该实验研究对于调节银纳米簇的荧光强度以及设计适配体为G四联体的荧光探针开关具有借鉴意义。 Abstract:Objective To investigate the effect of different guanine base numbers on the fluorescence intensity of DNA/ silver nanoclusters through C-G base complementary pairing, in order to explore a new method for the construction of fluorescent probe switches. Methods Designed complementary sequences of aptamer parts of a series of silver nanoclusters by using the nucleic acid base complementary pairing principle, and investigated the effect of the number of G bases exposed in the aptamer sequence on the fluorescence signal. Results Base G could enhance the fluorescence signal intensity of silver nanoclusters, and the fluorescence signal strength was positively correlated with the number of G bases. The fitting linear equation was Y=1726.1X+8972.5, r=0.9789. Conclusion This study is a great reference for the regulation of fluorescence intensity of silver nanoclusters and the design of G quadruplet aptamer fluorescent probe switch. -
Key words:
- DNA/AgNCs /
- complementary pairing /
- fluorescence probe /
- aptamer
-
紫苏为重要的药食两用中药,来源于唇形科植物紫苏Perilla frutescens(L.)Britt,干燥叶与成熟果实分别作为紫苏叶与紫苏子入药,历版《中国药典》均有收载,紫苏子能降气化痰,止咳平喘,润肠通便,用于痰壅气逆,咳嗽气喘,肠燥便秘[1]。紫苏叶有解表散寒,行气和胃的功效,用于风寒感冒,咳嗽呕恶,妊娠呕吐,鱼蟹中毒。
紫苏在我国栽培极广,紫苏叶除了供药用外,还可作蔬菜、香料以及代茶。紫苏叶和肉类煮熟可增加后者的香味。日本人多用于料理,作为生鱼片佐料。紫苏子榨出的油(苏子油)也可供食用。由于紫苏应用历史悠久,形态变异大,栽培品种多,通常将叶背腹面均绿色者称为白苏,叶背为紫色者称为紫苏。紫苏与白苏的分类处理至今仍存在较大争议。如《中国植物志》[2]与《中国药典》[1]均将紫苏与白苏合为一种。而《中药志》[3]《中药辞海》[4]《新编中药志》[5]以及《中药品种理论与应用》[6]等均认为紫苏为白苏的变种。
研究表明,紫苏与白苏的形态差别并非由于栽培条件差异导致,其化学成分方面也存在明显区别,紫苏的挥发油类成分主为紫苏醛型,而白苏为紫苏酮型[7],本文对药食两用中药紫苏的名称、形态和功效进行本草考证,以正本清源,为临床安全合理应用提供科学依据。
1. 紫苏与白苏的名称考证
紫苏原名苏,始载于《名医别录》,列为中品。李时珍在《本草纲目》中也记载:“苏性舒畅,行气和血,故谓之苏”[8]。陶弘景所著的《本草集经注》记载,“苏,味辛,温。主下气,除寒中,子尤良。叶下紫色而气甚香,其无紫色不香似荏者,名野苏,不堪用”。
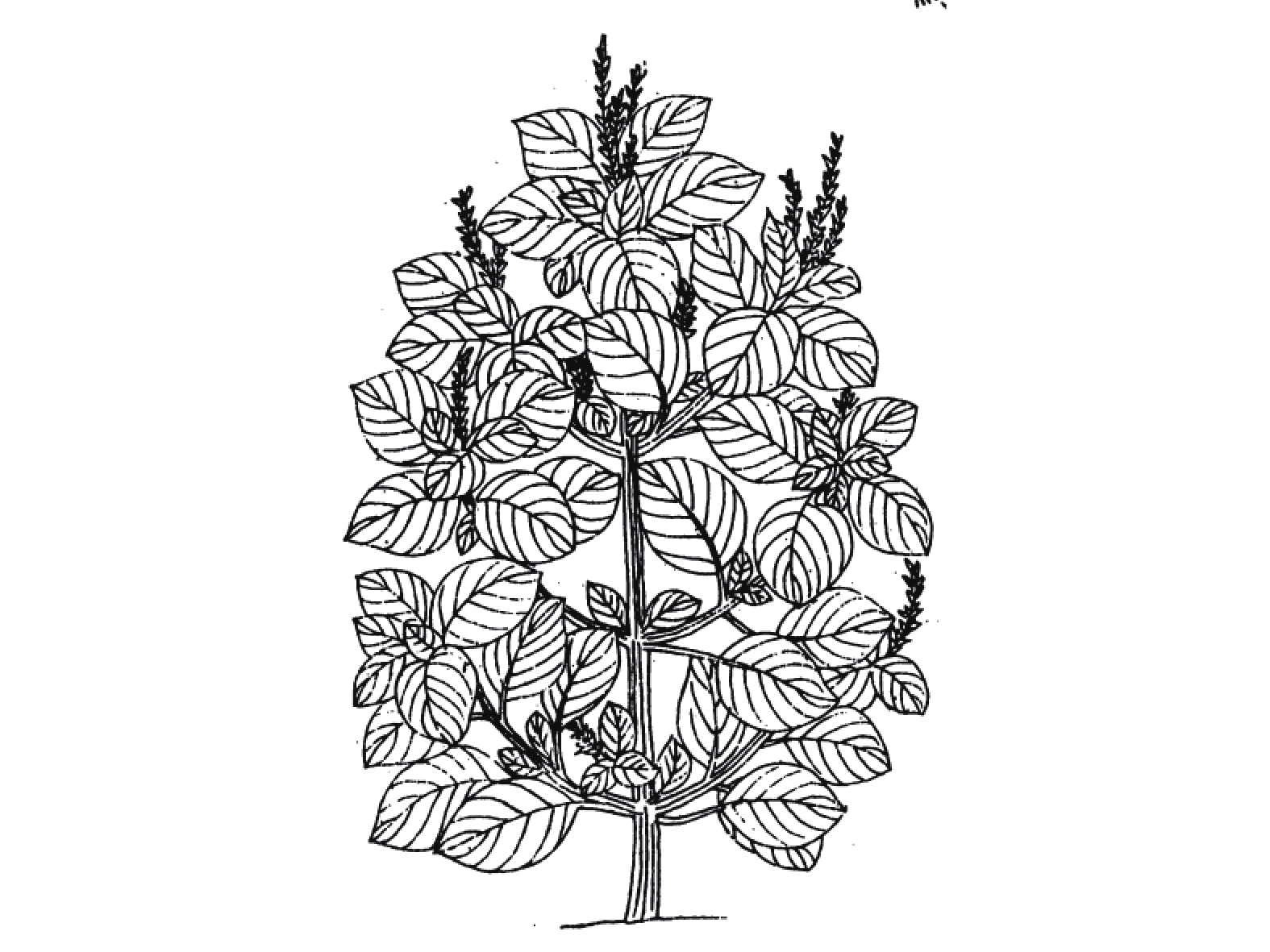
宋代苏颂所著的《图经本草》记载:“苏,紫苏也。谨按《尔雅》谓苏为桂荏。盖以其味辛,而形类荏,乃名之。然而苏有数种,有水苏、白苏、鱼苏、山鱼苏,皆是荏类”[9]。根据其形态描述及附图(图1),并为后来的本草文献支持,苏为现代唇形科植物紫苏Perilla frutescens (Linn.) Britt.。《救荒本草》记载:紫苏一名桂荏,又有数种,有勺苏、鱼苏、山苏,出简州及无为军,今处处有之[10]。这里提到的桂荏即紫苏,李时珍在《本草纲目》 解释到:“苏乃荏类,而味更辛如桂,故《尔雅 》谓之桂荏”。产地简州及无为军,《宋史》载:“无为军,同下州。太平兴国三年,以庐州巢县无为镇建为军,以巢、庐江二县来属。”《图经本草》有附图简州苏、无为军苏,与此也相符。
苏(舒)的名称来源于其功效。《尔雅义疏》也云:“苏之为言舒也”。紫苏则是指苏之茎叶之色为紫色。《本草求真》记载,紫苏专入肺,兼入心 、脾。背面俱紫,辛温香窜,凡风寒偶伤,气闭不利,心膨气胀,并暑湿泄泻,热闭血衄、崩淋,喉腥口臭,俱可用此调治。取其辛能入气,紫能入血,香能透外,温可暖中,使其一身舒畅,故命其名曰苏[11]。紫苏又称紫舒,相传还与东汉名医华佗有关。一次华佗在某地水边采药,无意中发现一只水獭因吃了螃蟹,难受在地上打滚,找到一种紫色的草吃了后竟安然无事。后来华佗用这种紫色的草熬汤救治了因多食螃蟹腹痛的患者。因该草紫色,患者喝后病痛解除,顿觉舒服,故称之为“紫舒”[12]。
古代本草中提到的“荏”即“白苏”,桂荏则指紫苏。李时珍《本草纲目》载:“曰紫苏者,以别白苏也。其面背皆白者即白苏,乃荏也”。《植物名实图考》中对荏也有解释,“荏,别录中品,白苏也”[13]。至于现代紫苏名称较多,紫苏又称野苏、红苏、香苏,青苏等名字。白苏又称白紫苏、青苏、臭苏等。
2. 紫苏与白苏的形态考证
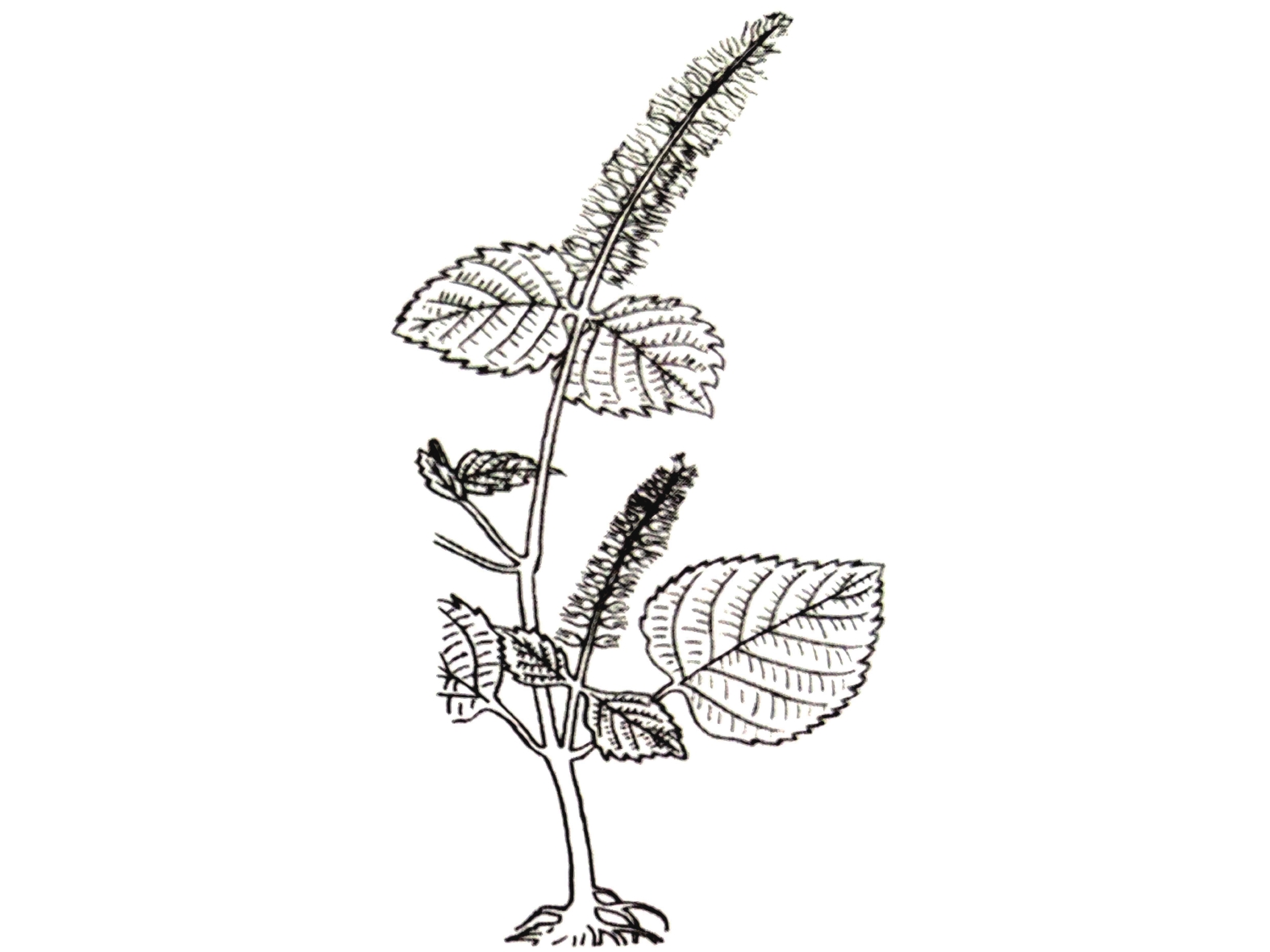
紫苏与白苏形态方面的差异目前争议颇多,分类上多被作为同一种处理,认为其形态的差异是由栽培条件不同导致。然而古代本草记载以及现代研究均表明紫苏、白苏在形态与成分方面存在明显差异。《图经本草》记载:“紫苏,叶下紫色,而气甚香,夏采茎叶,秋采实。《本草纲目》曰:“紫苏,其茎方,其叶圆而有尖,四围有锯齿;肥地者面背皆紫,瘠地者面青背紫”,并有附图(图2)。据此有人认为紫苏与白苏的形态差异由于生长环境导致。《本草崇原》也记载:紫苏,其叶面青背紫,气甚辛香,开花成穗,红紫色,穗中有细子,其色黄赤,入土易生,后人于壤土莳植,面背皆紫者名家紫苏。野生瘠土者,背紫面青。《别录》首次提到野生与栽培紫苏的区别。此外,还提到一种面背皆青,气辛臭香者,为荠。一种面背皆白者,名白苏,俱不堪入药。这里提到的“荠”应为现唇形科植物荠苧Mosla grosseserrata Maxim.,又称臭苏,青白苏。而《植物名实图考》记载紫苏并有附图(图3),形态特征十分清晰,与现代文献描述的紫苏形态特征相符。
白苏又称为荏,最大区别为叶不为紫色,历代本草对此均有记载,如《本草经集注》载:“荏,状如苏,高大白色,不甚香”;而《本草图经》载:“苏有数种,有水苏、白苏、鱼苏、山鱼苏,皆为荏类。白苏,方茎圆叶,不紫,亦甚香,实亦入药。事实上无论紫苏,白苏皆含挥发油,具有特殊香气,不过紫苏味更辛如桂。《本草纲目》 :“其面背皆白者即白苏,乃荏也”。《救荒本草》记载:“(荏)苗高一二尺,茎方。叶似薄荷叶,极肥大。开淡紫色花,结穗似紫苏穗,其子如黍,其枝茎对节生”(图4)。苏与荏形态上较为相似,主要为叶面颜色差异。
3. 紫苏与白苏的功效考证
紫苏叶、紫苏梗、紫苏子均可入药,紫苏叶具有解表散寒,行气和胃的功效,紫苏梗具有理气宽中,止痛,安胎的功效。紫苏子具有降气化痰,止咳平喘,润肠通便的功效。在历代本草中均有详细记载,如《别录》:“主下气,除寒中。”《食疗本草》:“除寒热,治冷气”[14] 。《日华子本草》:“补中益气。治心腹胀满,止霍乱转筋,开胃下食,并一切冷气,止脚气,通大小肠。”《履巉岩本草》:“止金疮出血,疗痔疾,煎汤洗之”。紫苏叶入药始见于南北朝时期的《雷公炮炙论》[15]。《图经本草》记载,其茎并叶,通心经,益脾胃,煮饮尤甚,与橘皮相宜,气方中多用之。 实主上气咳逆,……,若欲宣通风毒,则单用茎,去节良。
《滇南本草》:“苏叶,发汗,解伤风头疼,定吼喘,下气,宽臌,消胀、消痰。苏子,止咳嗽,降痰,定吼喘,下气,消痰涎。” 明代《本草汇言》则记载:“紫苏,散寒气,清肺气,宽中气,下结气,化痰气,乃治气之神药也”,突出强调其治气功效。明代《本草纲目》也记载:“(紫苏)行气宽中,消痰利肺,和血,温中,止痛,定喘,安胎。其味辛,入气分,其色紫,入血分。” 清代《本经逢原》则认为紫苏可“散血脉之邪。”[16]。
关于紫苏梗的功效,明代《医学入门·本草》记载:“(紫苏)治风寒湿痹,及筋骨疼痛,脚气” [17]。明代《本草通玄》认为其“能行气安胎”。清代《本草崇原》:“主宽中行气,消饮食,化痰涎。治噎膈反胃,止心腹痛”。清代《得配本草》记载:“疏肝,利肺,理气,和血,解郁,安胎” [18]。 明代《本草蒙筌》[19]:“下诸气略缓,体虚者用宜”。
清代张志聪的《侣山堂类辩》曾有详细记载[20]:“庭前植百合、紫苏各数茎,见百合花昼开夜合,紫苏叶朝挺暮垂,因悟草木之性,感天地阴阳之气而为开阖者也,……,苏色紫赤,枝茎空通,其气朝出暮入,有如经脉之气,昼行于阳,夜行于阴,是以苏叶能发表汗者,血液之汗也(白走气分,亦走血分)。枝茎能通血脉,故易思兰先生常用苏茎通十二经之关窍,治咽膈饱闷,通大小便,止下利赤白。予亦常用香苏细茎,不切断,治反胃膈食,吐血下血,多奏奇功”。详细描述了紫苏不同部位的功效,紫苏长于解表理气之功效,紫苏叶与紫苏茎功效各有不同。
紫苏与白苏均有食用记载,《证类本草》将紫苏列为菜部中品,将荏列为菜部上品[21]。紫苏多以叶食用,《救荒本草》记载,“紫苏一名桂荏,今处处有之,苗高二尺许,茎方叶似苏子叶微小,茎叶背面皆紫色而气甚……,救饥采叶炒食,煮饮亦可,子研汁煮粥食之皆好。叶可生食与鱼作羹味佳”。白苏的食用主要为种子,“子可炒食;又研杂米作粥,甚肥美。亦可笮油用”[10]。《食物本草》也记载:“(紫苏)子,研汁煮粥长食,令人肥白身香”[22]。紫苏食用沿袭至今,许多地区仍有紫苏叶生食并与生鱼片同食习惯。
紫苏子作药用,秦、汉时期《别录》就有记载:“主下气,除寒中”。南北朝《本草经集注》记载:“苏子,主下气,与橘皮相宜同疗也”。唐代《药性论》则有不同的记载,认为苏子“主上气咳逆,治冷气及腰脚中湿风结气”。五代时期《日华子本草》则认为紫苏子“主调中,益五脏,下气,止霍乱、呕吐、反胃,补虚劳,肥健人,利大小便,破癥结,消五膈,止嗽,润心肺,消痰气”。北宋时期《本草衍义》记载紫苏子“治肺气喘急”[23]。《本草纲目》认为“苏子与叶同功,发散风气宜用叶,清利上下则宜用子也”。
紫苏以种子、叶、梗入药,白苏多以种子、叶和根入药。白苏又称南苏,叶功效与紫苏相似,而白苏根又有洗疮祛风的功效。《别录》记载:“主治欬逆,下气,温中”。《日华子本草》记载:“调气,润心肺,长肌肤,益颜色,消宿食,止上气咳嗽,去狐臭,敷蛇咬”。《滇南本草》则记载:“南苏,治伤寒发热,无汗头痛,其效如神。此草治一切风寒,痰涌结而霍乱转筋,咳嗽吐痰、小儿风症,定痛止喘。梗能补中益气。根能洗疮祛风。子能开胃健脾”。
4. 紫苏的毒性与副作用
紫苏的毒副作用记载较少,多数不良反应为药物或食物的配伍导致。如《本草纲目》记载:“不可同鲤鱼食,生毒疮”。明代的《神农本草经疏》[24]:“病属阴虚,因发寒热或恶寒及头痛者,慎毋投之,以病宜敛宜补故也。火升作呕者,亦不宜服,惟可用子”。《本草通玄》则记载:“久服泄人真气”。《药性切用》提到“气虚者禁用”。《医学入门·本草》也提到“脾胃气虚常泄者禁用”。《本经逢原》记载:“性主疏泄,气虚久嗽,阴虚喘逆,脾虚便溏者皆不可用”。因紫苏性温,主下气,故阴虚、气虚及温病患者慎用。
我们对紫苏与白苏的形态与化学成分研究结果表明,白苏的挥发油类成分主要为紫苏酮型,具有一定的毒性,存在安全隐患,因此作为食用以紫苏为好,不宜使用白苏。
5. 小结
通过对紫苏名称的文献考证结果表明,历代本草所言“紫苏”“苏”“白苏”均属“荏”类,为唇形科紫苏Perilla frutescens及其变种,“紫苏”源于“苏”,为其形态与功效的延伸。李时珍区分“紫苏”与“白苏”,白苏又称为荏,紫苏又称桂荏。通常将叶背腹面均绿色者称为白苏,叶背为紫色称为紫苏。白苏不甚香,不堪入药。紫苏茎、叶、子均入药,主下气,治风寒。
根据历代本草记载,紫苏和白苏为两种不同的植物,现代分类处理尚存在争议,认为紫苏是同种植物因环境不同产生的变异。然而其形态、功效及化学成分均存在明显差别。根据历代草本的紫苏附图、形态描述,紫苏与白苏应作为不同的分类单元。
紫苏茎、叶、子均入药,功效有所不同,对于不同病症注意合理的配伍,以免产生不良反应。此外,气虚、阴虚及温病患者慎用。紫苏作为药食两用的种类,应用较为广泛,为保证临床应用的安全有效,应与白苏加以区分。
-
表 1 DNA/AgNCs的模板序列
名称 序列(5′—3′) P1C5 GGAGGTGGTGGGGCCCCCTAATTCCCCC P1AC5 GGAGGTGGTGGGGACCCCCTAATTCCCCC P1A5C5 GGAGGTGGTGGGGAAAAACCCCCTAATTCCCCC P1N GGAGGTGGTGGGGCCCTAACTCCCC P1Y GGAGGTGGTGGGGCCCTTAATCCCC 表 2 P1A5C5的互补序列
名称 序列(5′—3′) 0G CCTCCACCACCCCTTTTT 1G CTCCACCACCCCTTTTT 2G TCCACCACCCCTTTTT 4G TCCACCCCTTTTT 5G CACCCCTTTTT 6G ACCCCTTTTT 7G CCCTTTTT -
[1] CHEN X, KO S K, KIM M J, et al. A thiol-specific fluorescent probe and its application for bioimaging[J]. Chem Commun (Camb),2010,46(16):2751-2753. doi: 10.1039/b925453f [2] ZHAO J, GAO J, XUE W, et al. Upconversion luminescence-activated DNA nanodevice for ATP sensing in living cells[J]. J Am Chem Soc,2018,140(2):578-581. doi: 10.1021/jacs.7b11161 [3] AMIRI S, AHMADI R, SALIMI A, et al. Ultrasensitive and highly selective FRET aptasensor for Hg2+ measurement in fish samples using carbon dots/AuNPs as donor/acceptor platform[J]. New J Chem,2018,42(19):16027-16035. doi: 10.1039/C8NJ02781A [4] GUO H, LI J S, LI Y W, et al. A turn-on fluorescent sensor for Hg2+ detection based on graphene oxide and DNA aptamers[J]. New J Chem,2018,42(13):11147-11152. doi: 10.1039/C8NJ01709C [5] FANG B Y, LI C, AN J, et al. HIV-related DNA detection through switching on hybridized quenched fluorescent DNA-Ag nanoclusters[J]. Nanoscale,2018,10(12):5532-5538. doi: 10.1039/C7NR09647J [6] GUO W, YUAN J, DONG Q, et al. Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification[J]. J Am Chem Soc,2010,132(3):932-934. doi: 10.1021/ja907075s [7] WANG L J, REN M, ZHANG Q Y, et al. Excision repair-initiated enzyme-assisted bicyclic cascade signal amplification for ultrasensitive detection of uracil-DNA glycosylase[J]. Anal Chem,2017,89(8):4488-4494. doi: 10.1021/acs.analchem.6b04673 [8] XU N, WANG Q, LEI J, et al. Label-free triple-helix aptamer as sensing platform for “signal-on” fluorescent detection of thrombin[J]. Talanta,2015,132:387-391. doi: 10.1016/j.talanta.2014.09.031 [9] LATORRE A, SOMOZA Á. DNA-mediated silver nanoclusters: synthesis, properties and applications[J]. Chembiochem,2012,13(7):951-958. doi: 10.1002/cbic.201200053 [10] LIU J W. DNA-stabilized, fluorescent, metal nanoclusters for biosensor development[J]. Trac Trends Anal Chem,2014,58:99-111. doi: 10.1016/j.trac.2013.12.014 [11] YEH H C, SHARMA J, HAN J J, et al. A DNA-silver nanocluster probe that fluoresces upon hybridization[J]. Nano Lett,2010,10(8):3106-3110. doi: 10.1021/nl101773c [12] WALCZAK S, MORISHITA K, AHMED M, et al. Towards understanding of poly-guanine activated fluorescent silver nanoclusters[J]. Nanotechnology,2014,25(15):155501. doi: 10.1088/0957-4484/25/15/155501 [13] RICHARDS C I, CHOI S, HSIANG J C, et al. Oligonucleotide-stabilized Ag nanocluster fluorophores[J]. J Am Chem Soc,2008,130(15):5038-5039. doi: 10.1021/ja8005644 [14] MA J L, YIN B C, YE B C. DNA template-regulated intergrowth of a fluorescent silver nanocluster emitter pair[J]. RSC Adv,2015,5(119):98467-98471. doi: 10.1039/C5RA21159J [15] LIN R, TAO G, CHEN Y, et al. Constructing a robust fluorescent DNA-stabilized silver nanocluster probe module by attaching a duplex moiety[J]. Chemistry,2017,23(45):10893-10900. doi: 10.1002/chem.201701879 [16] JIANG Y T, TANG Y G, MIAO P. Polydopamine nanosphere@silver nanoclusters for fluorescence detection of multiplex tumor markers[J]. Nanoscale,2019,11(17):8119-8123. doi: 10.1039/C9NR01307E [17] GAO S, ZHENG X, HU B, et al. Enzyme-linked, aptamer-based, competitive biolayer interferometry biosensor for palytoxin[J]. Biosens Bioelectron,2017,89(pt 2):952-958. -





 下载:
下载:


 下载:
下载: