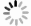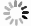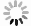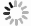-
由于免疫缺陷人群的增加和新真菌物种的出现,全球人口中侵袭性真菌感染(IFIs)越来越频繁[1-2]。IFIs对人类的发病率和死亡率有重大影响,全球每年造成约150万人死亡,并且这个数字还在增加[3-4]。
有研究发现,重症新型冠状病毒肺炎患者患侵袭性真菌感染的风险增加,有患者的临床相关标本中发现真菌标记物[5-6]。与非新冠肺炎患者相比,新冠肺炎患者中的侵袭性真菌感染发病率和死亡率都更高[7]。
目前临床常用的抗真菌药物有多烯类、唑类、棘白菌素和氟胞嘧啶等4类,其中氮唑类药物作为治疗和预防侵袭性真菌感染的一线疗法[8-9]。然而现有抗真菌药物的耐药性是一个日益严重的问题,具有可变敏感性或获得性耐药性的真菌种类数量呈增长趋势[3],且交叉耐药已被广泛报道[10]。有研究发现感染具有氟康唑和伏立康唑耐药念珠菌分离株患者的临床预后明显较差[11]。因此,开发具有高敏感的新型抗真菌药物迫在眉睫。
-
MSL300型核磁共振仪(CDCl3为溶剂,TMS为内标,Bruker公司);1260-1620LC-MS液相质谱联用仪、UPLC-QTOF/MS高分辨质谱仪(Agilent公司);HB10S096型旋转蒸发仪(IKA公司);2537型紫外分析仪(上海科艺光学仪器厂);DLSB型低温冷却液循环泵、B20-8-YK型耐腐蚀隔膜泵(上海豫康科教仪器设备有限公司)。监测反应使用的薄层色谱硅胶板(以硅胶GF254为固定相)和柱层析所用硅胶由烟台江友硅胶开发有限公司提供。起始原料(化合物1)、各类试剂均为市售分析纯或化学纯。
-
三唑类抗真菌药的作用机制是通过抑制对真菌细胞色素P450具有依赖性的羊毛甾醇14α-去甲基化酶,进而导致真菌细胞膜上麦角甾醇缺失,破坏真菌细胞立体结构的完整性,最终导致真菌的死亡[3,12]。根据国内外同行对三氮唑类化合物的研究,我们发现1,2,3-三唑是一种医学上有特殊意义的结构,在许多生物活性分子和药物中作为关键的结构特征。许多含有1,2,3-三唑结构的分子具有抗结核、抗真菌、抗过敏、抗病毒、抗肿瘤、抗疟疾作用以及神经保护活性[8,13-16]。
根据前人对氮唑类药物所作的构效关系的研究,其活性必需基团为叔醇结构和三唑环,2,4-二氟苯基为重要药效基团[17]。在此基础上,我们开展了以氟康唑为先导药物,设计合成新衍生物,引入不同的胺基侧链,尔后引入三氮唑环、各种取代苄基,观察活性情况。目标是获得高活性抗耐药的新型氮唑类化合物,丰富三氮唑类化合物的结构类型和构效关系。
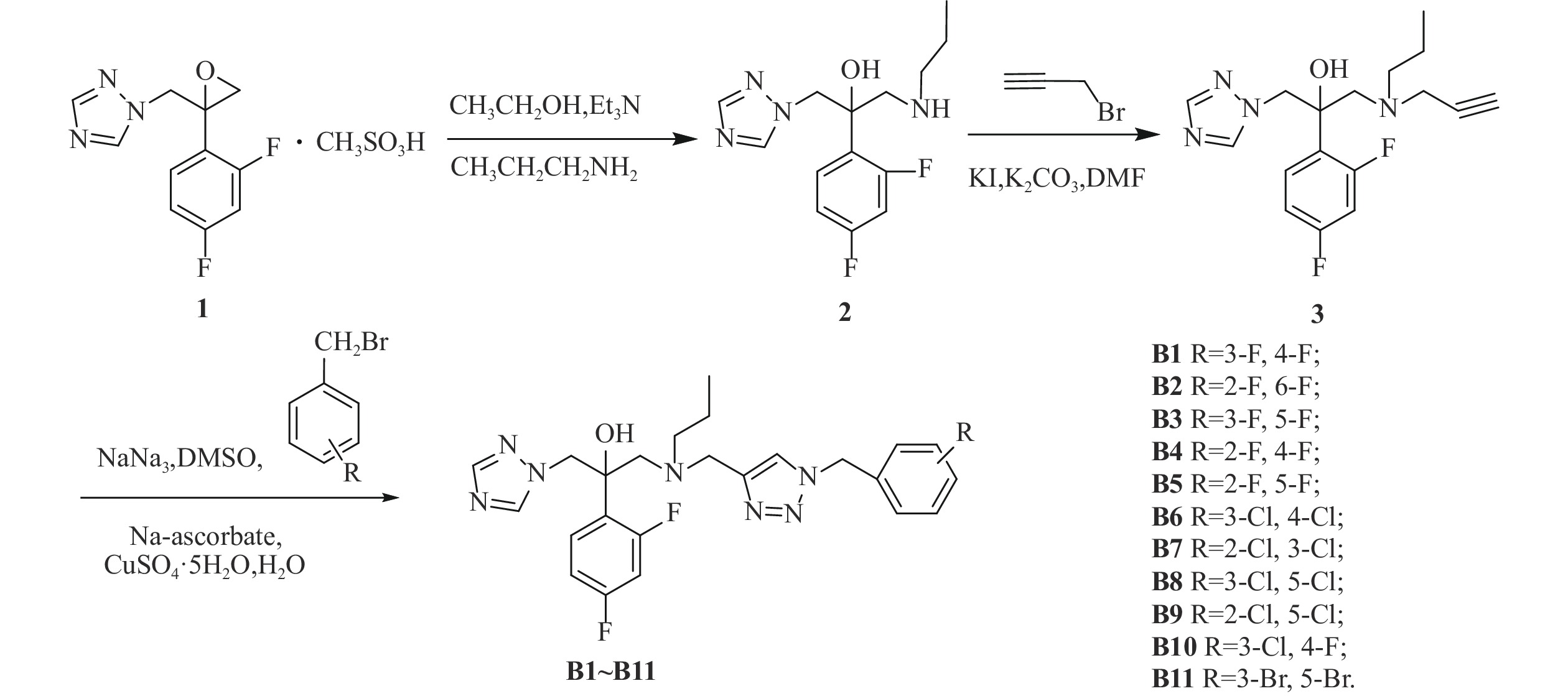
我们以1-[2-(2,4-二氟苯基)-2,3环氧丙基]-1H-1,2,4-三唑甲磺酸盐为起始原料,设计和合成了一系列氟康唑的类似物,结构为1-(1H-1,2,4-三唑-1-基)-2-(2,4-二氟苯基)-3-[N-正丙基-N-((1-取代-1H-1,2,3-三唑-4基)甲基)氨基]-2-丙醇类化合物。通过对3种人体致病真菌的初步抑菌试验,显示部分化合物具有一定的抗真菌活性。
目标化合物的合成路线如图1所示。
-
在500 ml的圆底烧瓶中加入化合物1(21 g, 63.05 mmol),正丙胺10 ml, 三乙胺20 ml,于250 ml乙醇中加热回流6~8 h[18],TLC监测,反应完毕后蒸除溶剂,用200 ml乙酸乙酯萃取,100 ml水洗2次,无水硫酸钠静置、干燥。过滤后旋蒸除掉乙酸乙酯,得油状的化合物2(12.042 g, 40.66 mmol),收率64.49%。
-
在250 ml的茄形瓶中加入化合物2(12.042 g, 40.66 mmol),溴丙炔(6.965 g, 58.55 mmol),无水碳酸钾(8.092 g, 58.55 mmol),碘化钾(675 mg, 4.066 mmol),于50 ml N,N-二甲基甲酰胺中室温下反应8 h[19],TLC监测,反应完毕后,用50 ml乙酸乙酯提取,100 ml水洗2次,无水硫酸钠静置、干燥。过滤后旋蒸除掉乙酸乙酯,柱层析[流动相为石油醚/乙酸乙酯(V/V)=1/1]得化合物3,收率76.27%。
-
在50 ml茄形瓶中加入叠氮钠(164 mg, 2.52 mmol)、3,4-二氟溴苄(261 mg, 1.26 mmol),二甲亚砜10 ml,室温下磁力搅拌反应3~5 h,然后加入化合物3(210 mg, 0.63 mmol),抗坏血酸钠 (20 mg, 0.10 mmol),无水硫酸铜 (25 mg, 0.10 mmol),水1 ml,室温搅拌反应10 min[19],TCL监测至化合物3反应完毕。将反应液倒入稀氨水中,乙酸乙酯萃取(30 ml×2次),乙酸乙酯层再用稀盐酸酸化萃取(30 ml×2次),分出水层,水层加无水碳酸钠调pH至7左右,乙酸乙酯萃取(30 ml×2次),乙酸乙酯层用无水硫酸钠干燥4 h后,过滤,减压抽溶剂得目标产物,收率67.99%。
-
方法同化合物B1,收率50.10%~95.83%。
中间体及目标化合物的波谱数据见表1, 部分化合物的高分辨质谱数据见表2。
表 1 目标化合物的结构和波谱数据
化合物 R 波谱数据 3 1H NMR(300 MHz, CDCl3) δ 8.14 (1H, s, triazole-H), 7.80 (1H, s, triazole-H), 7.59-7.50 (1H, q, Ar-H), 6.85-6.78 (2H, m, Ar-H), 5.06 (1H, s, -OH), 4.60-4.48 (2H, q, J = 11.35 Hz, –CH2), 3.32-3.27 (1H, dd, J=13.95 Hz, –CH2), 3.19-3.02 (2H, m, –CH2), 2.76-2.72 (1H, d, J = 13.96 Hz, –CH), 2.46-2.37 (1H, m, –CH2), 2.33-2.24 (1H, m, –CH2), 2.15 (1H, t, J=2.22 Hz, –CH), 1.34-1.22 (2H, m, –CH2), 0.79-0.74 (3H, t, J=7.35 Hz, –CH3).
13C NMR(75 MHz, CDCl3) δ 164.51-161.04, 160.69-157.27, 151.09, 144.68, 129.52, 125.97, 111.49, 104.30, 78.12, 73.06, 72.37, 58.43, 57.03, 56.19, 43.37, 20.61, 11.40.B1 3-F, 4-F 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, s, triazole-H), 7.77 (1H, s, triazole-H), 7.59 (1H, q, Ar-H), 7.18 (1H, d, triazole-H), 7.05 (3H, d, Ar-H), 6.80 (2H, m, Ar-H), 5.49 (2H, s, –CH2) , 4.56-4.51 (1H, d, J=13.80 Hz, –CH2), 4.01-4.36 (1H, d, J=13.71 Hz, –CH2), 3.65-3.49 (2H, q, J=16.28 Hz, –CH2), 3.19-3.14 (1H, d, J=13.24 Hz, –CH2), 2.73-2.68 (1H, d, J=13.90 Hz, –CH2), 2.27-2.17 (2H, q, J=29.82 Hz, –CH2), 1.29 (2H, s, –CH2), 0.70-0.65 (3H, t, J=6.78 Hz, –CH3). B2 2-F, 6-F 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, d, triazole-H), 7.75 (1H, s, triazole-H), 7.60-7.53 (1H, q, Ar-H), 7.42-7.32 (1H, m, Ar-H), 7.20 (1H, s, triazole-H), 7.00-6.95 (2H, t, Ar-H), 6.82-6.73 (2H, m, Ar-H), 5.61 (2H, s, –CH2), 4.51-4.38 (2H, q, J=13.55 Hz, –CH2), 3.61-3.46 (2H, q, J=14.43 Hz, –CH2), 3.17-3.13 (1H, d, J=14.20 Hz, –CH2), 2.73-2.69 (1H, d, J=13.63 Hz, –CH2), 2.24 (2H, s, –CH2), 1.27 (2H, s, –CH2), 0.70-0.65 (3H, t, J=6.98 Hz, –CH3). B3 3-F, 5-F 1H NMR(300 MHz, CDCl3) δ 8.08 (1H, d, triazole-H), 7.76 (1H, s, triazole-H), 7.63-7.55 (1H, m, Ar-H), 7.11 (1H, s, triazole-H), 6.85-6.74 (5H, m, Ar-H), 5.52 (2H, s, –CH2), 4.55-4.51 (1H, d, J=14.25 Hz, –CH2), 4.41-4.36 (1H, d, J=14.25 Hz, –CH2), 3.66-3.50 (2H, q, J=16.61 Hz, –CH2), 3.19-3.14 (1H, d, J=14.06 Hz, –CH2), 2.73-2.68 (1H, d, J=14.00 Hz, –CH2), 2.34-2.25 (1H, m, –CH2), 2.21-2.11 (1H, m, –CH2), 1.36-1.25 (2H, m, –CH2), 0.70-0.65 (3H, t, J=7.34 Hz, –CH3). B4 2-F, 4-F 1H NMR(300 MHz, CDCl3) δ 8.09 (1H, d, triazole-H), 7.76 (1H, s, triazole-H), 7.63-7.54 (1H, m, Ar-H), 7.31 (1H, t, Ar-H), 7.17 (1H, s, triazole-H), 6.91-6.85 (2H, t, Ar-H), 6.84-6.74 (2H, t, Ar-H), 5.53 (2H, s, –CH2), 4.54-4.37 (2H, q, J=17.08 Hz, –CH2), 3.58-3.53 (2H, d, J=15.43 Hz, –CH2), 3.18-3.14 (1H, d, J=11.92 Hz, –CH2), 2.74-2.71 (1H, d, J=9.30 Hz, –CH2), 2.27-2.20 (2H, d, J=20.71 Hz, –CH2), 1.29-1.25 (2H, d, J=12.28 Hz, –CH2), 0.70-0.66 (3H, t, J=6.27 Hz, –CH3). B5 2-F, 5-F 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, d, triazole-H), 7.77 (1H, s, triazole-H), 7.63-7.55 (1H, m, Ar-H), 7.20 (1H, s, triazole-H), 7.11-7.05 (2H, m, Ar-H), 6.94 (1H, s, Ar-H), 6.84-6.75 (2H, m, Ar-H), 5.56 (2H, s, –CH2), 4.55-4.39 (2H, q, J=16.18 Hz, –CH2), 3.64-3.50 (2H, d, J=13.86 Hz, –CH2), 3.20-3.15 (1H, d, J=15.01 Hz, –CH2), 2.75-2.71 (1H, d, J=13.51 Hz, –CH2), 2.29-2.22 (2H, d, J=21.24 Hz, –CH2), 1.30 (2H, s, –CH2), 0.69 (3H, s, –CH3). B6 3-Cl, 4-Cl 1H NMR(300 MHz, CDCl3) δ 8.11 (1H, d, triazole-H), 7.78 (1H, s, triazole-H), 7.63-7.55 (1H, m, Ar-H), 7.47-7.44 (1H, d, Ar-H), 7.35 (1H, s, triazole-H), 7.11-7.09 (2H, d, Ar-H), 6.84-6.75 (2H, m, Ar-H), 5.49 (2H, s, –CH2), 4.56-4.52 (1H, d, J=14.13 Hz, –CH2), 4.41-4.36 (1H, d, J=14.25 Hz, –CH2), 3.66-3.49 (2H, q, J=16.49 Hz, –CH2), 3.19-3.14 (1H, d, J=13.33 Hz, –CH2), 2.73-2.68 (1H, d, J=13.27 Hz, –CH2), 2.28-2.17 (2H, d, J=31.32 Hz, –CH2), 1.29 (2H, s, –CH2), 0.70-0.65 (3H, t, J=7.24 Hz, –CH3). B7 2-Cl, 3-Cl 1H NMR(300 MHz, CDCl3) δ 8.13 (1H, d, triazole-H), 7.77 (1H, s, triazole-H), 7.62-7.54 (1H, m, Ar-H), 7.50-7.47 (1H, dd, Ar-H), 7.25-7.19 (2H, t, Ar-H), 7.05 (1H, d, triazole-H), 6.84-6.73 (2H, m, Ar-H), 5.68 (2H, s, –CH2), 4.55-4.39 (2H, q, J=15.71 Hz, –CH2), 3.64-3.50 (2H, q, J=14.53 Hz, –CH2), 3.19-3.14 (1H, d, J=14.05 Hz, –CH2), 2.75-2.70 (1H, d, J=13.48 Hz, –CH2), 2.28-2.22 (2H, d, J=20.34 Hz, –CH2), 1.29 (2H, s, –CH2), 0.71-0.65 (3H, t, J=7.28 Hz, –CH3). B8 3-Cl, 5-Cl 1H NMR(300 MHz, CDCl3) δ 8.12 (1H, d, triazole-H), 7.78 (1H, s, triazole-H), 7.64-7.56 (1H, m, Ar-H), 7.36 (1H, s, triazole-H), 7.13 (3H, s, Ar-H), 6.86-6.75 (2H, m, Ar-H), 5.49 (2H, s, –CH2), 4.58-4.53 (1H, q, J=13.92 Hz, –CH2), 4.42-4.37 (1H, q, J=14.61 Hz, –CH2), 3.62-3.55 (2H, d, J=21.03 Hz, –CH2), 3.20-3.15 (1H, d, J=13.23 Hz, –CH2), 2.72-2.68 (1H, d, J=10.65 Hz, –CH2), 2.29-2.18 (2H, d, J=35.37 Hz, –CH2), 1.30 (2H, s, –CH2), 0.69 (3H, s, –CH3). B9 2-Cl, 5-Cl 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, d, triazole-H), 7.76 (1H, s, triazole-H), 7.63-7.54 (1H, m, Ar-H), 7.39-7.31 (2H, t, Ar-H), 7.21 (1H, s, triazole-H), 7.13 (1H, s, Ar-H), 6.84-6.75 (2H, m, Ar-H), 5.61 (2H, s, –CH2), 4.55-4.50 (1H, d, J=13.47 Hz, –CH2), 4.44-4.39 (1H, q, J=14.43 Hz, –CH2), 3.65-3.51 (2H, q, J=14.43 Hz, –CH2), 3.20-3.16 (1H, d, J=13.14 Hz, –CH2), 2.75-2.70 (1H, d, J=13.55 Hz, –CH2), 2.29-2.22 (2H, d, J=21.33 Hz, –CH2), 1.28 (2H, s, –CH2), 0.69 (3H, s, –CH3). B10 3-Cl, 4-F 1H NMR(300 MHz, CDCl3) δ 8.13 (1H, d, triazole-H), 7.78 (1H, s, triazole-H), 7.62-7.54 (1H, m, Ar-H), 7.32-7.30 (1H, d, Ar-H), 7.15-7.13 (2H, d, Ar-H), 7.09 (1H, s, triazole-H), 6.83-6.73 (2H, m, Ar-H), 5.47 (2H, s, –CH2), 4.55-4.36 (2H, q, J=19.44 Hz, –CH2), 3.65-3.48 (2H, q, J=16.33 Hz, –CH2), 3.18-3.13 (1H, d, J=13.91 Hz, –CH2), 2.72-2.68 (1H, d, J=13.77 Hz, –CH2), 2.33-2.24 (1H, m, –CH2), 2.20-2.10 (1H, m, –CH2), 1.29 (2H, s, –CH2), 0.69-0.64 (3H, t, J=7.31 Hz, –CH3).
13C NMR(75 MHz, CDCl3) δ 167.88-164.42, 164.02-160.59, 163.30, 159.98, 154.85, 148.29, 135.22, 133.72, 133.15, 131.26, 129.69, 125.38, 125.14, 120.71, 114.88, 107.58, 75.57, 61.42, 60.72, 59.63, 56.25, 52.75, 23.57, 14.81.B11 3-Br, 5-Br 1H NMR(300 MHz, CDCl3) δ 8.08 (1H, d, triazole-H), 7.77 (1H, s, triazole-H), 7.66 (1H, s, Ar-H), 7.63-7.55 (1H, m, Ar-H), 7.32 (2H, d, Ar-H), 7.10 (1H, d, triazole-H), 6.85-6.74 (2H, m, Ar-H), 5.47 (2H, s, –CH2), 5.33 (1H, s, -OH), 4.56-4.52 (1H, d, J=14.16 Hz, –CH2), 4.41-4.36 (1H, d, J=14.22 Hz, –CH2), 3.67-3.50 (2H, q, J=16.40 Hz, –CH2), 3.20-3.16 (1H, d, J=13.56 Hz, –CH2), 2.73-2.68 (1H, d, J=14.27 Hz, –CH2), 2.29-2.19 (2H, d, J=29.70 Hz, –CH2), 1.29 (2H, s, –CH2), 0.71-0.66 (3H, t, J=7.31 Hz, –CH3).
13C NMR(300 MHz, CDCl3) δ 164.50-161.03, 160.61-157.19, 150.88, 144.93, 138.40, 134.51, 129.75, 129.61, 129.61, 127.97, 126.35, 123.64, 123.64, 122.10, 111.51, 104.18, 72.23, 58.02, 57.36, 56.07, 52.66, 49.39, 20.18, 11.40.表 2 部分化合物的高分辨质谱数据
化合物 R ChemDraw提示分子量 HRMS测得分子量 3 334.16 335.1694 B2 2-F, 6-F 503.21 504.2127 B3 3-F, 5-F 503.21 504.2141 B5 2-F, 5-F 503.21 504.2152 B7 2-Cl, 3-Cl 535.15 536.1546 -
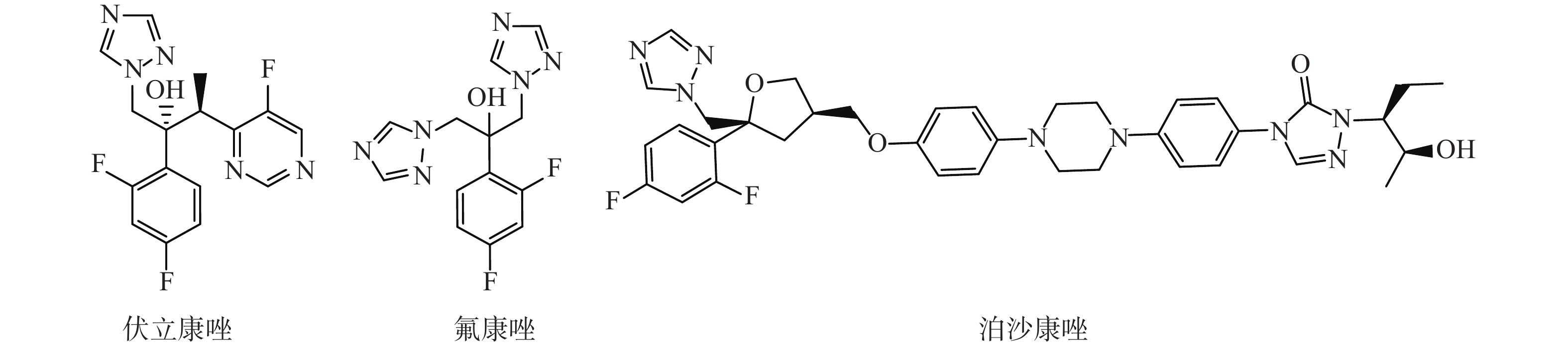
选取了3种实验真菌,菌株由海军军医大学药学系军特药研究中心提供,阳性对照药为伏立康唑(VCZ)、泊沙康唑(POS)和氟康唑(FCZ),对照药结构式见图2。
测试化合物体外抑菌活性的实验采用了美国国家临床实验室标准委员会(NCCLS)提出的标准化抗真菌敏感性实验方法。目标化合物对3种致病菌(C.alb SC5314, C.neo, A.fum.)的体外抑菌活性测试结果如表3所示。
表 3 目标化合物的体外抗真菌活性(MIC80, μg/ml)
化合物 R C.alb SC5314 C.neo h99 A.fum 7544 B1 3-F, 4-F 2 16 >64 B2 2-F, 6-F 1 8 >64 B3 3-F, 5-F 4 8 >64 B4 2-F, 4-F 1 4 >64 B5 2-F, 5-F 2 8 >64 B6 3-Cl, 4-Cl 2 8 >64 B7 2-Cl, 3-Cl 1 8 >64 B8 3-Cl, 5-Cl 4 >64 >64 B9 2-Cl, 5-Cl 1 16 >64 B10 3-Cl, 4-F 1 1 >64 B11 3-Br, 5-Br 0.125 2 >64 VCZ 0.0156 0.125 0.125 POS 0.125 0.5 1 FCZ 0.25 8 >64 注 :C.alb: 白念珠菌;C.neo: 新型隐球菌;A.fum: 烟曲霉菌;VCZ: 伏立康唑; POS: 泊沙康唑;FCZ: 氟康唑。 -
根据目标化合物的体外抗真菌活性结果,化合物B11对白念珠菌SC5314的活性与泊沙康唑相当,是氟康唑的2倍; 化合物B2、B3、B5、B6、B7对新型隐球菌H99的活性与氟康唑相当,化合物B10、B11、B4对新型隐球菌h99的活性是氟康唑的2~4倍;所有化合物对烟曲霉菌活性欠佳。总体来看,活性最好的3个化合物依次是化合物B11、B10、B4,R取代基分别为3-Br, 5-Br、3-Cl, 4-F、2-F、4-F,侧链中均含有正丙基,对除烟曲霉菌以外的其他2种真菌的抑制活性比较突出,分析可能是正丙基作为疏水基团与靶酶的疏水腔结合较好,苯基上间位的取代基减小了分子空间位阻,提高了化合物的抗真菌活性。由于本实验合成的化合物数量有限,更深入的构效关系探讨有待于进一步的研究。
Design, synthesis and antifungal activity of novel triazoles containing propyl side chains
-
摘要:
目的 研究具有正丙基侧链和二取代苯环结构的三唑醇类化合物的抗真菌活性。 方法 设计合成了11个目标化合物;其结构通过1H NMR确证,部分化合物还通过13C NMR、高分辨质谱(HRMS)确证;选择3种真菌为实验菌株,根据美国国家临床实验室标准委员会(NCCLS)推荐的标准化抗真菌敏感性实验方法,进行体外抑菌活性测试。 结果 化合物 B11 对白念珠菌SC5314的活性较氟康唑更好,与泊沙康唑相当;化合物 B10 、 B11 、 B4 对新型隐球菌H99的活性较氟康唑更好,化合物 B2 、 B3 、 B5 、 B6 、 B7 对新型隐球菌H99的活性与氟康唑相当;所有化合物对烟曲霉菌活性欠佳。 结论 部分引入正丙基侧链和二取代苯基结构的目标化合物有一定抗真菌活性,可作为潜在的先导抗真菌药物。 Abstract:Objective To study the antifungal activity of a new series of triazole compounds with n-propyl side chain and disubstituted benzyl structure. Methods Eleven target compounds were designed and synthesized. The structures were confirmed by 1H NMR, and some compounds were confirmed by 13C NMR or HRMS. Three fungal strains were selected as experimental strains, and the antifungal activity was tested in vitro according to the standardized antifungal sensitivity test method recommended by National Committee for Clinical Laboratory Standards (NCCLS). Results Compound B11 showed better activity against candida albicans SC5314 than fluconazole and was comparable to posaconazole; Compounds B10 , B11 and B4 showed better activity against cryptococcus neoformanis H99 than fluconazole, while compounds B2 , B3 , B5 , B6 and B7 showed similar activity to fluconazole against cryptococcus neoformanis H99; while all compounds showed poor activity against aspergillus fumigatus. Conclusion Some of the target compounds with n-propyl side chain and disubstituted benzyl group structure had certain antifungal activity and could be identified as potential lead antifungal drugs. -
糖尿病患者长期的高血糖环境显著影响自主神经系统,导致消化系统功能紊乱,表现为腹胀、便秘或腹泻等一系列消化不良的现象[1]。结肠镜检查作为评估结直肠疾病的关键工具,特别是在早期结肠癌的检测中扮演着不可或缺的角色。然而,不充分的肠道准备工作会直接影响检查的有效性。临床实践中,3 000 ml复方聚乙二醇电解质散剂被广泛采纳作为肠道准备的标准方案[2]。但对于糖尿病患者群体,一方面,自主神经损害减弱了胃肠道的正常蠕动;另一方面,部分患者接受的胰高血糖素样肽-1受体激动剂(GLP-1Ra)治疗进一步延缓了胃排空速度。在这两重因素共同作用下,糖尿病患者面临肠道清洁不佳的难题,严重影响了结肠镜检查的质量和病变检出率[3, 4]。胆宁片能改善消化过程,减少肠道负担,进一步促进排便[5],可显著优化糖尿病患者在接受结肠镜检查前的肠道准备状态,进而提升检查的准确性和舒适度。既往曾有研究报道胆宁片可改善2型糖尿病患者的便秘,但未见探讨其在肠道准备中的作用[6]。本研究旨在探索将胆宁片与复方聚乙二醇电解质散的预处理方案应用于糖尿病患者的结肠镜检查,评估胆宁片对提高肠道准备合格率的应用效果,现报告如下。
1. 资料与方法
1.1 一般资料
选取2022年11月至2024 年5月在海军军医大学第一附属医院内分泌科住院并行结肠镜检查的100例2型糖尿病患者为研究对象。根据是否使用GLP-1Ra类降糖药物(使用GLP-1Ra类药物共22名患者)进行分层随机,分为两组,每组各50例。所有研究对象均签署了知情同意书,本研究已通过我院伦理委员会批准。
纳入标准:①患者必须满足世界卫生组织(WHO)确立的2型糖尿病确诊标准;②研究对象的年龄≥30岁;③日常生活能力评估,即Barthel指数得分,须高于60分。④文化程度高中及以上。
排除标准:①糖尿病病情波动不定,且伴有如心脑血管突发事件等严重并发疾病的患者;②正处于糖尿病急性并发症发作期的患者;③患有急性胆囊炎、急性化脓性胆囊炎或任何急腹症状况、炎症性肠病、消化道梗阻、腹腔手术史、肝硬化、严重肾功能不全、帕金森病、心力衰竭的患者;④存在精神心理性疾病病史或近1周服三环类药物的患者;⑤过度肥胖(BMI>30)的患者[7-9]。
1.2 方法
1.2.1 对照组
①患者教育与心理准备:在肠道准备阶段之初,对参与者进行详尽的教育宣讲,着重说明肠道准备的必要性及其实施步骤,旨在减轻或消除患者面对结肠镜检查时可能产生的焦虑感,营造积极合作的氛围[10]。②膳食管理:要求受试者在结肠镜检查前1 d采用低残渣、半流质饮食模式,晚餐只可吃易消化的半流质食品,避免摄入富含纤维的食物。于检查当日清晨开始禁食,以减少肠道内容物,便于后续检查顺利进行[11]。③药物准备:在进行结肠镜检查前10 h,受试者应分次服用完68.56 g复方聚乙二醇电解质散(组成包括聚乙二醇4 000、无水硫酸钠、氯化钠、氯化钾和碳酸氢钠),药物需溶解在1 000 ml温水中。检查前6 h,受试者再次分次服用完137.12 g复方聚乙二醇电解质散,药物溶解在2 000 ml温水中,以确保肠道得到充分的清洁准备[12]。
1.2.2 实验组
在对照组的基础上从检查前2 d开始,口服胆宁片每日3次,每次5片,共服2 d,其余常规准备方案同对照组。
1.3 观察指标
肠道准备质量评估:采用波士顿量表(Boston Bowel Preparation Scale,BBPS),由肠镜室医生评估各组肠道准备质量。该量表将全结肠分为3个主要的结肠区域,分别为右半结肠(盲肠和升结肠)、横结肠(肝曲到脾曲的部分)、左半结肠(降结肠、乙状结肠和直肠)。每个区域的清洁度分数如下:0分:由于无法清除的固体或液体粪便导致整段肠黏膜无法观察;1分:由于污斑、混浊液体、残留粪便导致部分肠黏膜无法观察;2分:肠道黏膜观察良好,但残留少量污斑、混浊液体、粪便;3分:肠道黏膜观察良好,基本无残留污斑、混浊液体、粪便。每个区域的评分范围是从0~3分,总评分范围是0~9分,最终得分<6分为差,6~7分为良,8~9分为优。总评分越高表明肠道准备质量越好,有助于提高结肠镜检查的准确性和效率[13, 14]。
1.4 统计学方法
采用 SPSS 26.0 软件对数据进行统计分析,符合正态分布的计量资料以(均数±标准差)表示,组间比较用t检验;不符合正态分布的计量资料以中位数和四分位数区间表示,组间比较用非参数检验。计数资料(n, %)用卡方检验比较,以P<0.05 为差异有统计学意义。
2. 结果
2.1 一般资料
本研究共收集100名2型糖尿病患者,随机分为对照组和实验组,每组各50名,对照组和实验组各有11例患者使用GLP-1Ra类降糖药物。其中,对照组平均年龄(56.38±9.44)岁,男性占比72.0%(36/50),平均糖尿病病程(11.26±6.59)年,BMI为(24.21±1.76)kg/m2,糖化血红蛋白为(7.93±1.21)%;实验组平均年龄(57.42±9.99)岁,男性占比68.0%(34/50),平均病程(9.52±7.23)年,BMI为(24.31±2.47)kg/m2,糖化血红蛋白为(7.89±1.21)%。两组间基础资料对比,差异均无统计学意义(P>0.05)。
2.2 BBPS评价两组患者肠道准备质量
根据BBPS评分,评分为“优”的,实验组占66%,显著高于对照组的28%。评分为“良”的,两组都占30%。评分为“差”的,实验组仅4%,远低于对照组的42%。实验组平均分为(7.44±1.03),对照组为(5.58±1.98)。实验组在肠道准备方面表现明显优于对照组,且差异具有统计学意义(P<0.001),见表1。
表 1 BBPS评价肠道准备情况组别 评分 t值 P值 优 良 差 卡方值 P值 对照组 5.58±1.98 −0.589 <0.001 14
(28%)15
(30%)21
(42%)23.377 <0.001 实验组 7.44±1.03 33
(66%)15
(30%)2
(4%)2.3 BBPS评价两组使用GLP-1Ra药物的患者肠道准备质量
对照组和实验组各有11例患者使用GLP-1Ra类降糖药物,对使用GLP-1Ra的糖尿病患者单独进行比较:实验组的BBPS平均得分为(6.64±0.81),高于对照组的平均得分(3.64±0.92);实验组BBPS评分有1人被评为优,10人被评为良,而对照组全部为差。因两组样本量过小,无法进行统计学分析,但以上结果仍提示在使用了GLP-1Ra的情况下,实验组的肠道准备质量优于对照组。
3. 讨论
在进行结肠镜检查前的肠道准备中,糖尿病患者遇到的挑战尤为显著,这与他们复杂的生理病理特征密切相关。血糖水平对胃部活动和排空有直接调控作用。高血糖状态下,胃排空速度受到抑制,造成胃肠道的感觉与运动功能紊乱。病程较长的糖尿病患者可能存在神经病变,尤其是在消化系统方面导致一系列消化不良的症状,如腹胀、便秘或腹泻,影响胃肠道平滑肌的健康与功能,从而影响肠道准备的充分性[15]。
结肠镜检查对于评估结直肠疾病至关重要,尤其在早期结肠癌筛查中发挥关键作用。然而,若肠道准备不充分,可能会延长检查时间并增加并发症风险,直接影响检查的准确性和安全性[16]。尽管3 000 ml复方聚乙二醇电解质散剂是肠道准备的常用标准方案,但糖尿病患者因自主神经受损和使用GLP-1Ra药物治疗的影响,胃肠道蠕动减慢,胃排空延迟,使得肠道清洁难度增加,将直接影响结肠镜检查的效果[17]。
本研究结果显示,胆宁片能够显著改善糖尿病患者的肠道准备效果,尤其是在使用GLP-1Ra的患者中,作用更为明显。胆宁片是一种中药制剂,含有大黄、虎杖、青皮、陈皮、郁金和山楂等成分。山楂促进消化液分泌,增强胃肠蠕动,帮助消化油腻食物,同时还有一定的降脂作用;大黄具有较强的泻下作用,可以清除体内积热,改善便秘症状,并且有解毒、清热的效果;陈皮燥湿化痰,对脾胃功能不佳、消化不良有一定的辅助治疗效果;虎杖具有解毒、活血化瘀的功效,对于体内瘀血、炎症有一定的缓解作用;青皮疏肝理气,适用于肝气郁结引起的消化不良等症状。几种成分共同作用能有效清热、促进肠道运动、疏肝解郁、促进消化酶分泌、加速食物消化、减少肠道负担,同时增强胆汁分泌,帮助脂肪乳化,软化大便,利于排便[18]。
本研究同样面临一些局限性:第一,样本规模较为局限;第二,在探究糖尿病患者治疗方案时,未能细致分类伴随病症及所用降糖药物,此疏忽可能模糊了药物效能与不良反应情况;第三,缺乏对患者依从性的详细记录。未来仍需增加样本量、完善干预措施、评估相关药物以进一步验证和完善方案。
-
表 1 目标化合物的结构和波谱数据
化合物 R 波谱数据 3 1H NMR(300 MHz, CDCl3) δ 8.14 (1H, s, triazole-H), 7.80 (1H, s, triazole-H), 7.59-7.50 (1H, q, Ar-H), 6.85-6.78 (2H, m, Ar-H), 5.06 (1H, s, -OH), 4.60-4.48 (2H, q, J = 11.35 Hz, –CH2), 3.32-3.27 (1H, dd, J=13.95 Hz, –CH2), 3.19-3.02 (2H, m, –CH2), 2.76-2.72 (1H, d, J = 13.96 Hz, –CH), 2.46-2.37 (1H, m, –CH2), 2.33-2.24 (1H, m, –CH2), 2.15 (1H, t, J=2.22 Hz, –CH), 1.34-1.22 (2H, m, –CH2), 0.79-0.74 (3H, t, J=7.35 Hz, –CH3).
13C NMR(75 MHz, CDCl3) δ 164.51-161.04, 160.69-157.27, 151.09, 144.68, 129.52, 125.97, 111.49, 104.30, 78.12, 73.06, 72.37, 58.43, 57.03, 56.19, 43.37, 20.61, 11.40.B1 3-F, 4-F 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, s, triazole-H), 7.77 (1H, s, triazole-H), 7.59 (1H, q, Ar-H), 7.18 (1H, d, triazole-H), 7.05 (3H, d, Ar-H), 6.80 (2H, m, Ar-H), 5.49 (2H, s, –CH2) , 4.56-4.51 (1H, d, J=13.80 Hz, –CH2), 4.01-4.36 (1H, d, J=13.71 Hz, –CH2), 3.65-3.49 (2H, q, J=16.28 Hz, –CH2), 3.19-3.14 (1H, d, J=13.24 Hz, –CH2), 2.73-2.68 (1H, d, J=13.90 Hz, –CH2), 2.27-2.17 (2H, q, J=29.82 Hz, –CH2), 1.29 (2H, s, –CH2), 0.70-0.65 (3H, t, J=6.78 Hz, –CH3). B2 2-F, 6-F 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, d, triazole-H), 7.75 (1H, s, triazole-H), 7.60-7.53 (1H, q, Ar-H), 7.42-7.32 (1H, m, Ar-H), 7.20 (1H, s, triazole-H), 7.00-6.95 (2H, t, Ar-H), 6.82-6.73 (2H, m, Ar-H), 5.61 (2H, s, –CH2), 4.51-4.38 (2H, q, J=13.55 Hz, –CH2), 3.61-3.46 (2H, q, J=14.43 Hz, –CH2), 3.17-3.13 (1H, d, J=14.20 Hz, –CH2), 2.73-2.69 (1H, d, J=13.63 Hz, –CH2), 2.24 (2H, s, –CH2), 1.27 (2H, s, –CH2), 0.70-0.65 (3H, t, J=6.98 Hz, –CH3). B3 3-F, 5-F 1H NMR(300 MHz, CDCl3) δ 8.08 (1H, d, triazole-H), 7.76 (1H, s, triazole-H), 7.63-7.55 (1H, m, Ar-H), 7.11 (1H, s, triazole-H), 6.85-6.74 (5H, m, Ar-H), 5.52 (2H, s, –CH2), 4.55-4.51 (1H, d, J=14.25 Hz, –CH2), 4.41-4.36 (1H, d, J=14.25 Hz, –CH2), 3.66-3.50 (2H, q, J=16.61 Hz, –CH2), 3.19-3.14 (1H, d, J=14.06 Hz, –CH2), 2.73-2.68 (1H, d, J=14.00 Hz, –CH2), 2.34-2.25 (1H, m, –CH2), 2.21-2.11 (1H, m, –CH2), 1.36-1.25 (2H, m, –CH2), 0.70-0.65 (3H, t, J=7.34 Hz, –CH3). B4 2-F, 4-F 1H NMR(300 MHz, CDCl3) δ 8.09 (1H, d, triazole-H), 7.76 (1H, s, triazole-H), 7.63-7.54 (1H, m, Ar-H), 7.31 (1H, t, Ar-H), 7.17 (1H, s, triazole-H), 6.91-6.85 (2H, t, Ar-H), 6.84-6.74 (2H, t, Ar-H), 5.53 (2H, s, –CH2), 4.54-4.37 (2H, q, J=17.08 Hz, –CH2), 3.58-3.53 (2H, d, J=15.43 Hz, –CH2), 3.18-3.14 (1H, d, J=11.92 Hz, –CH2), 2.74-2.71 (1H, d, J=9.30 Hz, –CH2), 2.27-2.20 (2H, d, J=20.71 Hz, –CH2), 1.29-1.25 (2H, d, J=12.28 Hz, –CH2), 0.70-0.66 (3H, t, J=6.27 Hz, –CH3). B5 2-F, 5-F 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, d, triazole-H), 7.77 (1H, s, triazole-H), 7.63-7.55 (1H, m, Ar-H), 7.20 (1H, s, triazole-H), 7.11-7.05 (2H, m, Ar-H), 6.94 (1H, s, Ar-H), 6.84-6.75 (2H, m, Ar-H), 5.56 (2H, s, –CH2), 4.55-4.39 (2H, q, J=16.18 Hz, –CH2), 3.64-3.50 (2H, d, J=13.86 Hz, –CH2), 3.20-3.15 (1H, d, J=15.01 Hz, –CH2), 2.75-2.71 (1H, d, J=13.51 Hz, –CH2), 2.29-2.22 (2H, d, J=21.24 Hz, –CH2), 1.30 (2H, s, –CH2), 0.69 (3H, s, –CH3). B6 3-Cl, 4-Cl 1H NMR(300 MHz, CDCl3) δ 8.11 (1H, d, triazole-H), 7.78 (1H, s, triazole-H), 7.63-7.55 (1H, m, Ar-H), 7.47-7.44 (1H, d, Ar-H), 7.35 (1H, s, triazole-H), 7.11-7.09 (2H, d, Ar-H), 6.84-6.75 (2H, m, Ar-H), 5.49 (2H, s, –CH2), 4.56-4.52 (1H, d, J=14.13 Hz, –CH2), 4.41-4.36 (1H, d, J=14.25 Hz, –CH2), 3.66-3.49 (2H, q, J=16.49 Hz, –CH2), 3.19-3.14 (1H, d, J=13.33 Hz, –CH2), 2.73-2.68 (1H, d, J=13.27 Hz, –CH2), 2.28-2.17 (2H, d, J=31.32 Hz, –CH2), 1.29 (2H, s, –CH2), 0.70-0.65 (3H, t, J=7.24 Hz, –CH3). B7 2-Cl, 3-Cl 1H NMR(300 MHz, CDCl3) δ 8.13 (1H, d, triazole-H), 7.77 (1H, s, triazole-H), 7.62-7.54 (1H, m, Ar-H), 7.50-7.47 (1H, dd, Ar-H), 7.25-7.19 (2H, t, Ar-H), 7.05 (1H, d, triazole-H), 6.84-6.73 (2H, m, Ar-H), 5.68 (2H, s, –CH2), 4.55-4.39 (2H, q, J=15.71 Hz, –CH2), 3.64-3.50 (2H, q, J=14.53 Hz, –CH2), 3.19-3.14 (1H, d, J=14.05 Hz, –CH2), 2.75-2.70 (1H, d, J=13.48 Hz, –CH2), 2.28-2.22 (2H, d, J=20.34 Hz, –CH2), 1.29 (2H, s, –CH2), 0.71-0.65 (3H, t, J=7.28 Hz, –CH3). B8 3-Cl, 5-Cl 1H NMR(300 MHz, CDCl3) δ 8.12 (1H, d, triazole-H), 7.78 (1H, s, triazole-H), 7.64-7.56 (1H, m, Ar-H), 7.36 (1H, s, triazole-H), 7.13 (3H, s, Ar-H), 6.86-6.75 (2H, m, Ar-H), 5.49 (2H, s, –CH2), 4.58-4.53 (1H, q, J=13.92 Hz, –CH2), 4.42-4.37 (1H, q, J=14.61 Hz, –CH2), 3.62-3.55 (2H, d, J=21.03 Hz, –CH2), 3.20-3.15 (1H, d, J=13.23 Hz, –CH2), 2.72-2.68 (1H, d, J=10.65 Hz, –CH2), 2.29-2.18 (2H, d, J=35.37 Hz, –CH2), 1.30 (2H, s, –CH2), 0.69 (3H, s, –CH3). B9 2-Cl, 5-Cl 1H NMR(300 MHz, CDCl3) δ 8.10 (1H, d, triazole-H), 7.76 (1H, s, triazole-H), 7.63-7.54 (1H, m, Ar-H), 7.39-7.31 (2H, t, Ar-H), 7.21 (1H, s, triazole-H), 7.13 (1H, s, Ar-H), 6.84-6.75 (2H, m, Ar-H), 5.61 (2H, s, –CH2), 4.55-4.50 (1H, d, J=13.47 Hz, –CH2), 4.44-4.39 (1H, q, J=14.43 Hz, –CH2), 3.65-3.51 (2H, q, J=14.43 Hz, –CH2), 3.20-3.16 (1H, d, J=13.14 Hz, –CH2), 2.75-2.70 (1H, d, J=13.55 Hz, –CH2), 2.29-2.22 (2H, d, J=21.33 Hz, –CH2), 1.28 (2H, s, –CH2), 0.69 (3H, s, –CH3). B10 3-Cl, 4-F 1H NMR(300 MHz, CDCl3) δ 8.13 (1H, d, triazole-H), 7.78 (1H, s, triazole-H), 7.62-7.54 (1H, m, Ar-H), 7.32-7.30 (1H, d, Ar-H), 7.15-7.13 (2H, d, Ar-H), 7.09 (1H, s, triazole-H), 6.83-6.73 (2H, m, Ar-H), 5.47 (2H, s, –CH2), 4.55-4.36 (2H, q, J=19.44 Hz, –CH2), 3.65-3.48 (2H, q, J=16.33 Hz, –CH2), 3.18-3.13 (1H, d, J=13.91 Hz, –CH2), 2.72-2.68 (1H, d, J=13.77 Hz, –CH2), 2.33-2.24 (1H, m, –CH2), 2.20-2.10 (1H, m, –CH2), 1.29 (2H, s, –CH2), 0.69-0.64 (3H, t, J=7.31 Hz, –CH3).
13C NMR(75 MHz, CDCl3) δ 167.88-164.42, 164.02-160.59, 163.30, 159.98, 154.85, 148.29, 135.22, 133.72, 133.15, 131.26, 129.69, 125.38, 125.14, 120.71, 114.88, 107.58, 75.57, 61.42, 60.72, 59.63, 56.25, 52.75, 23.57, 14.81.B11 3-Br, 5-Br 1H NMR(300 MHz, CDCl3) δ 8.08 (1H, d, triazole-H), 7.77 (1H, s, triazole-H), 7.66 (1H, s, Ar-H), 7.63-7.55 (1H, m, Ar-H), 7.32 (2H, d, Ar-H), 7.10 (1H, d, triazole-H), 6.85-6.74 (2H, m, Ar-H), 5.47 (2H, s, –CH2), 5.33 (1H, s, -OH), 4.56-4.52 (1H, d, J=14.16 Hz, –CH2), 4.41-4.36 (1H, d, J=14.22 Hz, –CH2), 3.67-3.50 (2H, q, J=16.40 Hz, –CH2), 3.20-3.16 (1H, d, J=13.56 Hz, –CH2), 2.73-2.68 (1H, d, J=14.27 Hz, –CH2), 2.29-2.19 (2H, d, J=29.70 Hz, –CH2), 1.29 (2H, s, –CH2), 0.71-0.66 (3H, t, J=7.31 Hz, –CH3).
13C NMR(300 MHz, CDCl3) δ 164.50-161.03, 160.61-157.19, 150.88, 144.93, 138.40, 134.51, 129.75, 129.61, 129.61, 127.97, 126.35, 123.64, 123.64, 122.10, 111.51, 104.18, 72.23, 58.02, 57.36, 56.07, 52.66, 49.39, 20.18, 11.40.表 2 部分化合物的高分辨质谱数据
化合物 R ChemDraw提示分子量 HRMS测得分子量 3 334.16 335.1694 B2 2-F, 6-F 503.21 504.2127 B3 3-F, 5-F 503.21 504.2141 B5 2-F, 5-F 503.21 504.2152 B7 2-Cl, 3-Cl 535.15 536.1546 表 3 目标化合物的体外抗真菌活性(MIC80, μg/ml)
化合物 R C.alb SC5314 C.neo h99 A.fum 7544 B1 3-F, 4-F 2 16 >64 B2 2-F, 6-F 1 8 >64 B3 3-F, 5-F 4 8 >64 B4 2-F, 4-F 1 4 >64 B5 2-F, 5-F 2 8 >64 B6 3-Cl, 4-Cl 2 8 >64 B7 2-Cl, 3-Cl 1 8 >64 B8 3-Cl, 5-Cl 4 >64 >64 B9 2-Cl, 5-Cl 1 16 >64 B10 3-Cl, 4-F 1 1 >64 B11 3-Br, 5-Br 0.125 2 >64 VCZ 0.0156 0.125 0.125 POS 0.125 0.5 1 FCZ 0.25 8 >64 注 :C.alb: 白念珠菌;C.neo: 新型隐球菌;A.fum: 烟曲霉菌;VCZ: 伏立康唑; POS: 泊沙康唑;FCZ: 氟康唑。 -
[1] GUPTA A K, VENKATARAMAN M, RENAUD H J, et al. The increasing problem of treatment-resistant fungal infections: a call for antifungal stewardship programs[J]. Int J Dermatol,2021,60(12):e474-e479. [2] CLARK C, DRUMMOND R A. The hidden cost of modern medical interventions: how medical advances have shaped the prevalence of human fungal disease[J]. Pathogens,2019,8(2):45. doi: 10.3390/pathogens8020045 [3] VAN DAELE R, SPRIET I, WAUTERS J, et al. Antifungal drugs: what brings the future? Med Mycol,2019,57(Supplement_3):S328-S343. doi: 10.1093/mmy/myz012 [4] BROWN G D, DENNING D W, GOW N A R, et al. Hidden killers: human fungal infections[J]. Sci Transl Med,2012,4(165):165rv13. [5] PEMÁN J, RUIZ-GAITÁN A, GARCÍA-VIDAL C, et al. Fungal co-infection in COVID-19 patients: should we be concerned? Rev Iberoam Micol,2020,37(2):41-46. doi: 10.1016/j.riam.2020.07.001 [6] PAPADIMITRIOU-OLIVGERIS M, KOLONITSIOU F, KEFALA S, et al. Increased incidence of candidemia in critically ill patients during the Coronavirus Disease 2019 (COVID-19) pandemic[J]. Braz J Infect Dis,2022,26(2):102353. doi: 10.1016/j.bjid.2022.102353 [7] SHISHIDO A A, MATHEW M, BADDLEY J W. Overview of COVID-19-associated invasive fungal infection[J]. Curr Fungal Infect Rep,2022,16(3):87-97. doi: 10.1007/s12281-022-00434-0 [8] CAO X F, WANG W D, WANG S S, et al. Asymmetric synthesis of novel triazole derivatives and their in vitro antiviral activity and mechanism of action[J]. Eur J Med Chem,2017,139:718-725. doi: 10.1016/j.ejmech.2017.08.057 [9] 元子青云, 陈安九, 沈怡雯, 等. 三唑类抗真菌药物临床应用研究进展[J]. 药学与临床研究, 2018, 26(2):125-129. doi: 10.13664/j.cnki.pcr.2018.02.013 [10] AGUILAR G, DELGADO C, CORRALES I, et al. Epidemiology of invasive candidiasis in a surgical intensive care unit: an observational study[J]. BMC Res Notes,2015,8:491. doi: 10.1186/s13104-015-1458-4 [11] PFALLER M A. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment[J]. Am J Med, 2012, 125(1 Suppl): S3-S13. [12] SANATI H, BELANGER P, FRATTI R, et al. A new triazole, voriconazole (UK-109, 496), blocks sterol biosynthesis in Candida albicans and Candida krusei[J]. Antimicrob Agents Chemother,1997,41(11):2492-2496. doi: 10.1128/AAC.41.11.2492 [13] WANG X L, WAN K, ZHOU C H. Synthesis of novel sulfanilamide-derived 1, 2, 3-triazoles and their evaluation for antibacterial and antifungal activities[J]. Eur J Med Chem,2010,45(10):4631-4639. doi: 10.1016/j.ejmech.2010.07.031 [14] LI J C, ZHANG J, RODRIGUES M C, et al. Synthesis and evaluation of novel 1, 2, 3-triazole-based acetylcholinesterase inhibitors with neuroprotective activity[J]. Bioorg Med Chem Lett,2016,26(16):3881-3885. doi: 10.1016/j.bmcl.2016.07.017 [15] BOECHAT N, FERREIRA V F, FERREIRA S B, et al. Novel 1, 2, 3-triazole derivatives for use against Mycobacterium tuberculosis H37Rv (ATCC 27294) strain[J]. J Med Chem,2011,54(17):5988-5999. doi: 10.1021/jm2003624 [16] THIRUMURUGAN P, MATOSIUK D, JOZWIAK K. Click chemistry for drug development and diverse chemical-biology applications[J]. Chem Rev,2013,113(7):4905-4979. doi: 10.1021/cr200409f [17] DOGAN S, SARAÇ S, SARI S, et al. New azole derivatives showing antimicrobial effects and their mechanism of antifungal activity by molecular modeling studies[J]. Eur J Med Chem,2017,130:124-138. doi: 10.1016/j.ejmech.2017.02.035 [18] QIAN A R, ZHENG Y Z, WANG R L, et al. Design, synthesis, and structure-activity relationship studies of novel tetrazole antifungal agents with potent activity, broad antifungal spectrum and high selectivity[J]. Bioorg Med Chem Lett,2018,28(3):344-350. doi: 10.1016/j.bmcl.2017.12.040 [19] RASTEGARI A, NADRI H, MAHDAVI M, et al. Design, synthesis and anti-Alzheimer's activity of novel 1, 2, 3-triazole-chromenone carboxamide derivatives[J]. Bioorg Chem,2019,83:391-401. doi: 10.1016/j.bioorg.2018.10.065 -




 下载:
下载:
 下载:
下载: